5 Tidyverse R
Hadley Wickham and Garrett Grolemund, in their excellent and freely available book R for Data Science, promote the concept of “tidy data.” The Tidyverse collection of R packages attempt to realize this concept in concrete libraries.
In brief, tidy data carefully separates variables (the columns of a table, also called features or fields) from observations (the rows of a table, also called samples). At the intersection of these two, we find values, one data item (datum) in each cell. Unfortunately, the data we encounter is often not arranged in this useful way, and it requires normalization. In particular, what are really values are often represented either as columns or as rows instead. To demonstrate what this means, let us consider an example (a small elementary school class).
library(tidyverse)
# inline reading, tibble version
students <- tribble(
~'Last Name', ~'First Name', ~'4th Grade', ~'5th Grade', ~'6th Grade',
"Johnson", "Mia", "A", "B+", "A-",
"Lopez", "Liam", "B", "B", "A+",
"Lee", "Isabella", "C", "C-", "B-",
"Fisher", "Mason", "B", "B-", "C+",
"Gupta", "Olivia", "B", "A+", "A",
"Robinson", "Sophia", "A+", "B-", "A"
)
students
#> # A tibble: 6 x 5
#> `Last Name` `First Name` `4th Grade` `5th Grade`
#> <chr> <chr> <chr> <chr>
#> 1 Johnson Mia A B+
#> 2 Lopez Liam B B
#> 3 Lee Isabella C C-
#> 4 Fisher Mason B B-
#> 5 Gupta Olivia B A+
#> 6 Robinson Sophia A+ B-
#> # ... with 1 more variable: 6th Grade <chr>This view of the data is easy for humans to read. We can see trends in the scores each student received over several years of education. Moreover, this format might lend itself to useful visualizations fairly easily:
# Generic conversion of letter grades to numbers
recodes.str <- "'A+'=4.3;'A'=4;'A-'=3.7;'B+'=3.3;'B'=3;'B-'=2.7;'C+'=2.3;'C'= 2;'C-'=1.7"
students$`4th Grade` <- car::recode(students$`4th Grade`, recodes.str)
students$`5th Grade` <- car::recode(students$`5th Grade`, recodes.str)
students$`6th Grade` <- car::recode(students$`6th Grade`, recodes.str)
# create plot
matplot(t(students[,c(3:5)]), type = "b", pch = 11:16, col = 2:7, xaxt="n", ylab="")
Axis(labels = names(students)[3:5], side=1, at = 1:3)
legend(1.2, 3, paste(substr(x = students$`Last Name`, 1, 1), students$`First Name`, sep = ". "),
pch = 11:16, col = 2:7)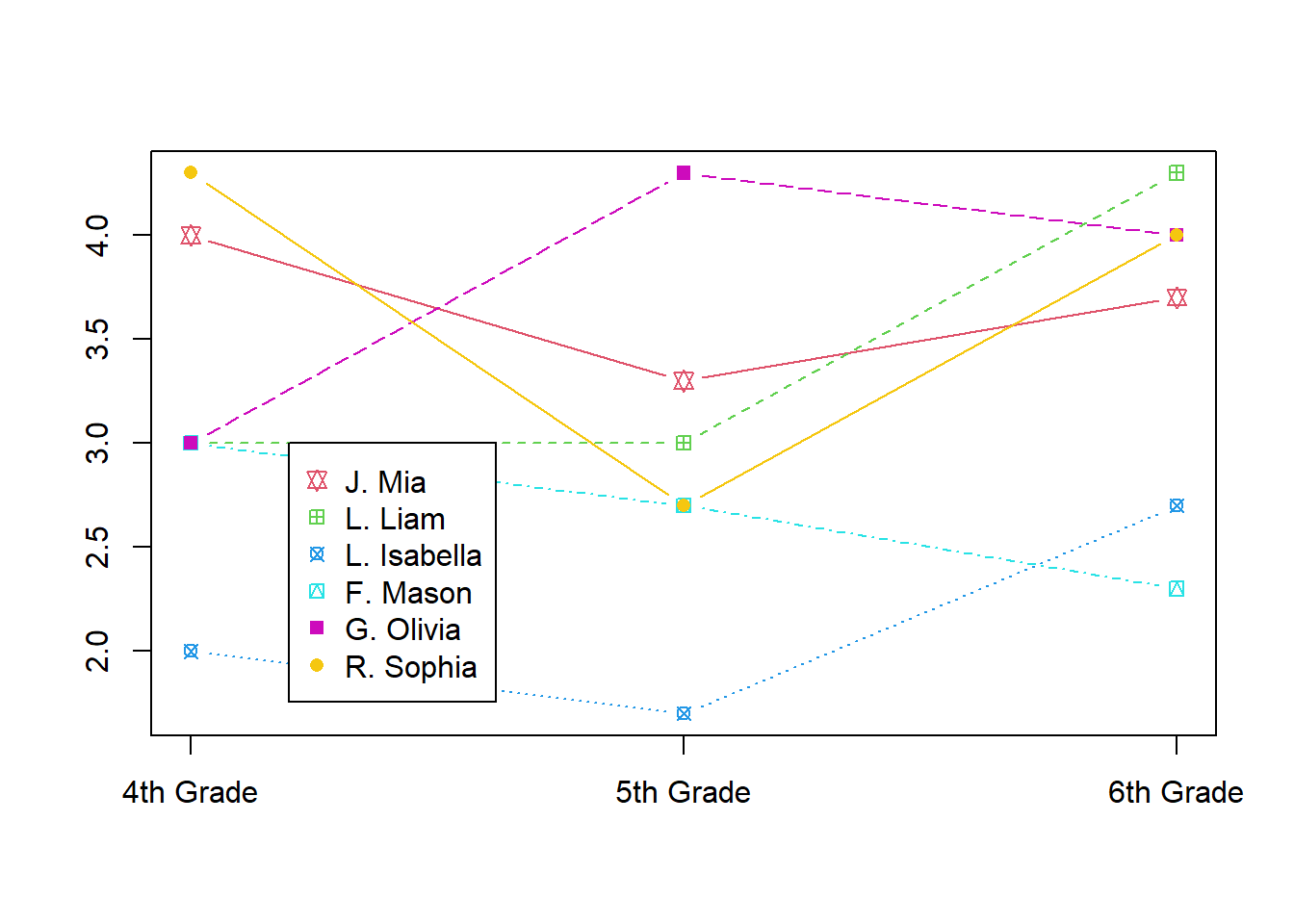
This data layout exposes its limitations once the class advances to 7th grade, or if we were to obtain 3rd grade information. To accommodate such additional data, we would need to change the number and position of columns, not simply add additional rows. It is natural to make new observations or identify new samples (rows) but usually awkward to change the underlying variables (columns).
The particular class level (e.g. 4th grade) that a letter grade pertains to is, at heart, a value, not a variable. Another way to think of this is in terms of independent variables versus dependent variables, or in machine learning terms, features versus target. In some ways, the class level might correlate with or influence the resulting letter grade; perhaps the teachers at the different levels have different biases, or children of a certain age lose or gain interest in schoolwork, for example.
For most analytic purposes, this data would be more useful if we made it tidy (normalized) before further processing. In Base R, the reshape2::melt() method can perform this tidying. We pin some of the columns as id_vars, and we set a name for the combined columns as a variable and the letter grade as a single new column.
reshape2::melt(data = students, id=c("Last Name", "First Name"))
#> Last Name First Name variable value
#> 1 Johnson Mia 4th Grade 4.0
#> 2 Lopez Liam 4th Grade 3.0
#> 3 Lee Isabella 4th Grade 2.0
#> 4 Fisher Mason 4th Grade 3.0
#> 5 Gupta Olivia 4th Grade 3.0
#> 6 Robinson Sophia 4th Grade 4.3
#> 7 Johnson Mia 5th Grade 3.3
#> 8 Lopez Liam 5th Grade 3.0
#> 9 Lee Isabella 5th Grade 1.7
#> 10 Fisher Mason 5th Grade 2.7
#> 11 Gupta Olivia 5th Grade 4.3
#> 12 Robinson Sophia 5th Grade 2.7
#> 13 Johnson Mia 6th Grade 3.7
#> 14 Lopez Liam 6th Grade 4.3
#> 15 Lee Isabella 6th Grade 2.7
#> 16 Fisher Mason 6th Grade 2.3
#> 17 Gupta Olivia 6th Grade 4.0
#> 18 Robinson Sophia 6th Grade 4.0Within the Tidyverse, specifically within the tidyr package, there is a function pivot_longer() that is similar to Base R’s reshape2::melt(). The aggregation names and values have parameters spelled names_to= and values_to=, but the operation is the same:
s.l <- students %>%
pivot_longer(c('4th Grade', '5th Grade', '6th Grade'),
names_to = "Level",
values_to = "Score")
s.l
#> # A tibble: 18 x 4
#> `Last Name` `First Name` Level Score
#> <chr> <chr> <chr> <dbl>
#> 1 Johnson Mia 4th Grade 4
#> 2 Johnson Mia 5th Grade 3.3
#> 3 Johnson Mia 6th Grade 3.7
#> 4 Lopez Liam 4th Grade 3
#> 5 Lopez Liam 5th Grade 3
#> 6 Lopez Liam 6th Grade 4.3
#> 7 Lee Isabella 4th Grade 2
#> 8 Lee Isabella 5th Grade 1.7
#> 9 Lee Isabella 6th Grade 2.7
#> 10 Fisher Mason 4th Grade 3
#> 11 Fisher Mason 5th Grade 2.7
#> 12 Fisher Mason 6th Grade 2.3
#> 13 Gupta Olivia 4th Grade 3
#> 14 Gupta Olivia 5th Grade 4.3
#> 15 Gupta Olivia 6th Grade 4
#> 16 Robinson Sophia 4th Grade 4.3
#> 17 Robinson Sophia 5th Grade 2.7
#> 18 Robinson Sophia 6th Grade 4The simple example above gives you a first feel for tidying tabular data. To reverse the tidying operation that moves variables (columns) to values (rows), the pivot_wider() function in tidyr can be used. In Base R there are several related methods on data frames, including reshape::cast() and reshape2::dcast().
s.l %>%
pivot_wider(names_from = Level, values_from = Score)
#> # A tibble: 6 x 5
#> `Last Name` `First Name` `4th Grade` `5th Grade`
#> <chr> <chr> <dbl> <dbl>
#> 1 Johnson Mia 4 3.3
#> 2 Lopez Liam 3 3
#> 3 Lee Isabella 2 1.7
#> 4 Fisher Mason 3 2.7
#> 5 Gupta Olivia 3 4.3
#> 6 Robinson Sophia 4.3 2.7
#> # ... with 1 more variable: 6th Grade <dbl>5.1 The tibble
Tibbles inherits the attributes of a data frame and enhances some of them. The tibble is the central data structure for a set of packages known as the tidyverse.
Tibbles when printed out returns:
- the first 10 rows and
- all the columns that can fit on screen and
- column types.
5.1.1 Importing data
The functions read_csv(), read_delim(), read_excel_csv(), read_tsv() are used to import data.
# loading package
library(readr)
# reading data
gapminder <- read_delim(file = 'data/gapminder_ext_UTF-8.txt',
delim = "\t",
col_names = T,
locale = locale(decimal_mark = ",", encoding = "UTF-8"))
head(gapminder, 3)
#> # A tibble: 3 x 8
#> country continent year lifeExp pop gdpPercap
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> # ... with 2 more variables: country_hun <chr>,
#> # continent_hun <chr>
# class checking
class(gapminder)
#> [1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"
# checking for data frame
is.data.frame(gapminder)
#> [1] TRUE5.1.2 Tibbles are data frames
Since Tibbles are data frames, functions which operate on data frames also operate on them.
head(gapminder, 3)
#> # A tibble: 3 x 8
#> country continent year lifeExp pop gdpPercap
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> # ... with 2 more variables: country_hun <chr>,
#> # continent_hun <chr>
tail(gapminder, 3)
#> # A tibble: 3 x 8
#> country continent year lifeExp pop gdpPercap
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Zimbabwe Africa 1997 46.8 11404948 792.
#> 2 Zimbabwe Africa 2002 40.0 11926563 672.
#> 3 Zimbabwe Africa 2007 43.5 12311143 470.
#> # ... with 2 more variables: country_hun <chr>,
#> # continent_hun <chr>
nrow(gapminder)
#> [1] 1704
ncol(gapminder)
#> [1] 8
summary(gapminder)
#> country continent year
#> Length:1704 Length:1704 Min. :1952
#> Class :character Class :character 1st Qu.:1966
#> Mode :character Mode :character Median :1980
#> Mean :1980
#> 3rd Qu.:1993
#> Max. :2007
#> lifeExp pop gdpPercap
#> Min. :23.60 Min. :6.001e+04 Min. : 241.2
#> 1st Qu.:48.20 1st Qu.:2.794e+06 1st Qu.: 1202.1
#> Median :60.71 Median :7.024e+06 Median : 3531.8
#> Mean :59.47 Mean :2.960e+07 Mean : 7215.3
#> 3rd Qu.:70.85 3rd Qu.:1.959e+07 3rd Qu.: 9325.5
#> Max. :82.60 Max. :1.319e+09 Max. :113523.1
#> country_hun continent_hun
#> Length:1704 Length:1704
#> Class :character Class :character
#> Mode :character Mode :character
#>
#>
#> 5.1.3 Exporting data
The functions write_csv(), write_delim(), write_excel_csv(), write_tsv() are used to export data. To export Tibbles, they have first to be converted into data frames.
# exporting Tibbles
write_delim(x = data.frame(gapminder), delim = " ", file = 'output/data/gapminderfixedwidth.txt')
write_csv(x = data.frame(gapminder), file = 'output/data/gapminder_csv.txt')
write_tsv(x = data.frame(gapminder), file = 'output/data/gapminder_tsv.txt')
# checking if files exist?
file.exists(c('output/data/gapminderfixedwidth.txt', 'output/data/gapminder_csv.txt', 'output/data/gapminder_tsv.txt'))
#> [1] TRUE TRUE TRUE
# removing files
file.remove(c('output/data/gapminderfixedwidth.txt', 'output/data/gapminder_csv.txt', 'output/data/gapminder_tsv.txt'))
#> [1] TRUE TRUE TRUE5.1.4 Check for tibble
Tibbles come from the package tibble.
The function is_tibble() is used to check for tibble.
The function glimpse() is a better option of str().
# loading tibble
library(tibble)
# glimpse() a better option to str()
glimpse(gapminder)
#> Rows: 1,704
#> Columns: 8
#> $ country <chr> "Afghanistan", "Afghanistan", "Afgha~
#> $ continent <chr> "Asia", "Asia", "Asia", "Asia", "Asi~
#> $ year <dbl> 1952, 1957, 1962, 1967, 1972, 1977, ~
#> $ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.0~
#> $ pop <dbl> 8425333, 9240934, 10267083, 11537966~
#> $ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.19~
#> $ country_hun <chr> "Afganisztán", "Afganisztán", "Afgan~
#> $ continent_hun <chr> "Ázsia", "Ázsia", "Ázsia", "Ázsia", ~
# checking whether an object is a tibble
is_tibble(gapminder)
#> [1] TRUE5.1.5 Creating a tibble
The function tibble() is like data.frame() but creates a tibble.
# creating named vectors
country <- c('China', 'India', 'United States', 'Indonesia', 'Brazil',
'Pakistan', 'Bangladesh', 'Nigeria', 'Japan', 'Mexico')
continent <- c('Asia', 'Asia', 'Americas', 'Asia', 'Americas',
'Asia', 'Asia', 'Africa', 'Asia', 'Americas')
population <- c(1318683096, 1110396331, 301139947, 223547000, 190010647,
169270617, 150448339, 135031164, 127467972, 108700891)
lifeExpectancy <- c(72.961, 64.698, 78.242, 70.65, 72.39,
65.483, 64.062, 46.859, 82.603, 76.195)
percapita <- c(4959, 2452, 42952, 3541, 9066, 2606, 1391, 2014, 31656, 11978)
# creating a tibble from named vectors
top_10 <- tibble(country, population, lifeExpectancy)
head(top_10, 3)
#> # A tibble: 3 x 3
#> country population lifeExpectancy
#> <chr> <dbl> <dbl>
#> 1 China 1318683096 73.0
#> 2 India 1110396331 64.7
#> 3 United States 301139947 78.2
class(top_10)
#> [1] "tbl_df" "tbl" "data.frame"5.1.6 Adding columns
The function add_column() is used to add columns to a tibble or data frames.
# adding a column to a tibble
# defaults to the last column
add_column(top_10, continent)
#> # A tibble: 10 x 4
#> country population lifeExpectancy continent
#> <chr> <dbl> <dbl> <chr>
#> 1 China 1318683096 73.0 Asia
#> 2 India 1110396331 64.7 Asia
#> 3 United States 301139947 78.2 Americas
#> 4 Indonesia 223547000 70.6 Asia
#> 5 Brazil 190010647 72.4 Americas
#> 6 Pakistan 169270617 65.5 Asia
#> 7 Bangladesh 150448339 64.1 Asia
#> 8 Nigeria 135031164 46.9 Africa
#> 9 Japan 127467972 82.6 Asia
#> 10 Mexico 108700891 76.2 Americas
# also works for data frames
add_column(as.data.frame(top_10), continent)
#> country population lifeExpectancy continent
#> 1 China 1318683096 72.961 Asia
#> 2 India 1110396331 64.698 Asia
#> 3 United States 301139947 78.242 Americas
#> 4 Indonesia 223547000 70.650 Asia
#> 5 Brazil 190010647 72.390 Americas
#> 6 Pakistan 169270617 65.483 Asia
#> 7 Bangladesh 150448339 64.062 Asia
#> 8 Nigeria 135031164 46.859 Africa
#> 9 Japan 127467972 82.603 Asia
#> 10 Mexico 108700891 76.195 Americas
# adding multiple columns
add_column(top_10, continent, percapita)
#> # A tibble: 10 x 5
#> country population lifeExpectancy continent percapita
#> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 China 1318683096 73.0 Asia 4959
#> 2 India 1110396331 64.7 Asia 2452
#> 3 United States 301139947 78.2 Americas 42952
#> 4 Indonesia 223547000 70.6 Asia 3541
#> 5 Brazil 190010647 72.4 Americas 9066
#> 6 Pakistan 169270617 65.5 Asia 2606
#> 7 Bangladesh 150448339 64.1 Asia 1391
#> 8 Nigeria 135031164 46.9 Africa 2014
#> 9 Japan 127467972 82.6 Asia 31656
#> 10 Mexico 108700891 76.2 Americas 11978
# adding multiple columns directly
add_column(top_10,
continent = c('Asia', 'Asia', 'Americas', 'Asia', 'Americas',
'Asia', 'Asia', 'Africa', 'Asia', 'Americas'),
percapita = c(4959, 2452, 42952, 3541, 9066, 2606, 1391, 2014, 31656, 11978))
#> # A tibble: 10 x 5
#> country population lifeExpectancy continent percapita
#> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 China 1318683096 73.0 Asia 4959
#> 2 India 1110396331 64.7 Asia 2452
#> 3 United States 301139947 78.2 Americas 42952
#> 4 Indonesia 223547000 70.6 Asia 3541
#> 5 Brazil 190010647 72.4 Americas 9066
#> 6 Pakistan 169270617 65.5 Asia 2606
#> 7 Bangladesh 150448339 64.1 Asia 1391
#> 8 Nigeria 135031164 46.9 Africa 2014
#> 9 Japan 127467972 82.6 Asia 31656
#> 10 Mexico 108700891 76.2 Americas 11978
# add a column before an index position
add_column(top_10, continent, .before = 2)
#> # A tibble: 10 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
#> 6 Pakistan Asia 169270617 65.5
#> 7 Bangladesh Asia 150448339 64.1
#> 8 Nigeria Africa 135031164 46.9
#> 9 Japan Asia 127467972 82.6
#> 10 Mexico Americas 108700891 76.2
# add a column after an index position
top_10 <- add_column(top_10, continent, .after = 1)
top_10
#> # A tibble: 10 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
#> 6 Pakistan Asia 169270617 65.5
#> 7 Bangladesh Asia 150448339 64.1
#> 8 Nigeria Africa 135031164 46.9
#> 9 Japan Asia 127467972 82.6
#> 10 Mexico Americas 108700891 76.25.1.7 Adding rows
The function add_row() is used to add rows to a tibble or a data frame.
# adding a row
# defaults to the tail of the data frame
add_row(top_10,
country = 'Philippines',
continent = 'Asia',
population = 91077287,
lifeExpectancy = 71.688)
#> # A tibble: 11 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
#> 6 Pakistan Asia 169270617 65.5
#> 7 Bangladesh Asia 150448339 64.1
#> 8 Nigeria Africa 135031164 46.9
#> 9 Japan Asia 127467972 82.6
#> 10 Mexico Americas 108700891 76.2
#> 11 Philippines Asia 91077287 71.7
# adding rows before an index position
add_row(top_10,
country = 'Philippines',
continent = 'Asia',
population = 91077287,
lifeExpectancy = 71.688,
.before = 2)
#> # A tibble: 11 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 Philippines Asia 91077287 71.7
#> 3 India Asia 1110396331 64.7
#> 4 United States Americas 301139947 78.2
#> 5 Indonesia Asia 223547000 70.6
#> 6 Brazil Americas 190010647 72.4
#> 7 Pakistan Asia 169270617 65.5
#> 8 Bangladesh Asia 150448339 64.1
#> 9 Nigeria Africa 135031164 46.9
#> 10 Japan Asia 127467972 82.6
#> 11 Mexico Americas 108700891 76.2
# adding rows after an index position
add_row(top_10,
country = 'Philippines',
continent = 'Asia',
population = 91077287,
lifeExpectancy = 71.688,
.after = 2)
#> # A tibble: 11 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 Philippines Asia 91077287 71.7
#> 4 United States Americas 301139947 78.2
#> 5 Indonesia Asia 223547000 70.6
#> 6 Brazil Americas 190010647 72.4
#> 7 Pakistan Asia 169270617 65.5
#> 8 Bangladesh Asia 150448339 64.1
#> 9 Nigeria Africa 135031164 46.9
#> 10 Japan Asia 127467972 82.6
#> 11 Mexico Americas 108700891 76.2
# adding multiple rows
add_row(top_10,
country = c('Philippines', 'Vietnam', 'Germany', 'Egypt', 'Ethiopia',
'Turkey', 'Iran', 'Thailand', 'Congo, Dem. Rep.', 'France'),
continent = c('Asia', 'Asia', 'Europe', 'Africa', 'Africa',
'Europe', 'Asia', 'Asia', 'Africa', 'Europe'),
population = c(91077287, 85262356, 82400996, 80264543, 76511887,
71158647, 69453570, 65068149, 64606759, 61083916),
lifeExpectancy = c(71.688, 74.249, 79.406, 71.338, 52.947,
71.777, 70.964, 70.616, 46.462, 80.657)
)
#> # A tibble: 20 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
#> 6 Pakistan Asia 169270617 65.5
#> 7 Bangladesh Asia 150448339 64.1
#> 8 Nigeria Africa 135031164 46.9
#> 9 Japan Asia 127467972 82.6
#> 10 Mexico Americas 108700891 76.2
#> 11 Philippines Asia 91077287 71.7
#> 12 Vietnam Asia 85262356 74.2
#> 13 Germany Europe 82400996 79.4
#> 14 Egypt Africa 80264543 71.3
#> 15 Ethiopia Africa 76511887 52.9
#> 16 Turkey Europe 71158647 71.8
#> 17 Iran Asia 69453570 71.0
#> 18 Thailand Asia 65068149 70.6
#> 19 Congo, Dem. Rep. Africa 64606759 46.5
#> 20 France Europe 61083916 80.75.1.8 Converting to tibble
The function as_tibble() is used to convert to a tibble, if possible.
# creating a matrix
mat = matrix(seq(1,12), 3, 4,
dimnames = list('a' = c('a1', 'a2', 'a3'), 'b' = c('b1', 'b2', 'b3', 'b4')))
mat
#> b
#> a b1 b2 b3 b4
#> a1 1 4 7 10
#> a2 2 5 8 11
#> a3 3 6 9 12
# converting a matrix to tibble
# removes the rownames
mat_tbl <- as_tibble(mat)
mat_tbl
#> # A tibble: 3 x 4
#> b1 b2 b3 b4
#> <int> <int> <int> <int>
#> 1 1 4 7 10
#> 2 2 5 8 11
#> 3 3 6 9 12
class(mat_tbl)
#> [1] "tbl_df" "tbl" "data.frame"
# creating a data frame
top_10_df <- data.frame(
country = c('China', 'India', 'United States', 'Indonesia', 'Brazil',
'Pakistan', 'Bangladesh', 'Nigeria', 'Japan', 'Mexico'),
continent = c('Asia', 'Asia', 'Americas', 'Asia', 'Americas',
'Asia', 'Asia', 'Africa', 'Asia', 'Americas'),
population = c(1318683096, 1110396331, 301139947, 223547000, 190010647,
169270617, 150448339, 135031164, 127467972, 108700891),
lifeExpectancy = c(72.961, 64.698, 78.242, 70.65, 72.39,
65.483, 64.062, 46.859, 82.603, 76.195)
)
head(top_10_df, 3)
#> country continent population lifeExpectancy
#> 1 China Asia 1318683096 72.961
#> 2 India Asia 1110396331 64.698
#> 3 United States Americas 301139947 78.242
class(top_10_df)
#> [1] "data.frame"
# converting data frame to tibble
top_tbl <- as_tibble(top_10_df)
top_tbl
#> # A tibble: 10 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
#> 6 Pakistan Asia 169270617 65.5
#> 7 Bangladesh Asia 150448339 64.1
#> 8 Nigeria Africa 135031164 46.9
#> 9 Japan Asia 127467972 82.6
#> 10 Mexico Americas 108700891 76.2
class(top_tbl)
#> [1] "tbl_df" "tbl" "data.frame"5.1.9 Manipulating row names
Tibble does not support row names but the package tibble has the following functions for dealing with row names:
-
has_rownames()checks if a data frame has row names. -
remove_rownames()removes row names. -
column_to_rownames()moves a column to row names. -
rowid_to_column()moves a row index to column.
# creating a data frame
top_10_df <- data.frame(
continent = c('Asia', 'Asia', 'Americas', 'Asia', 'Americas',
'Asia', 'Asia', 'Africa', 'Asia', 'Americas'),
population = c(1318683096, 1110396331, 301139947, 223547000, 190010647,
169270617, 150448339, 135031164, 127467972, 108700891),
lifeExpectancy = c(72.961, 64.698, 78.242, 70.65, 72.39,
65.483, 64.062, 46.859, 82.603, 76.195)
)
top_10_df
#> continent population lifeExpectancy
#> 1 Asia 1318683096 72.961
#> 2 Asia 1110396331 64.698
#> 3 Americas 301139947 78.242
#> 4 Asia 223547000 70.650
#> 5 Americas 190010647 72.390
#> 6 Asia 169270617 65.483
#> 7 Asia 150448339 64.062
#> 8 Africa 135031164 46.859
#> 9 Asia 127467972 82.603
#> 10 Americas 108700891 76.195
# vector of country names
country <- c('China', 'India', 'United States', 'Indonesia', 'Brazil',
'Pakistan', 'Bangladesh', 'Nigeria', 'Japan', 'Mexico')
# adding row names
rownames(top_10_df) <- country
top_10_df
#> continent population lifeExpectancy
#> China Asia 1318683096 72.961
#> India Asia 1110396331 64.698
#> United States Americas 301139947 78.242
#> Indonesia Asia 223547000 70.650
#> Brazil Americas 190010647 72.390
#> Pakistan Asia 169270617 65.483
#> Bangladesh Asia 150448339 64.062
#> Nigeria Africa 135031164 46.859
#> Japan Asia 127467972 82.603
#> Mexico Americas 108700891 76.195
# check if the data frame contains row names
has_rownames(top_10_df)
#> [1] TRUE
# delete row names
remove_rownames(top_10_df)
#> continent population lifeExpectancy
#> 1 Asia 1318683096 72.961
#> 2 Asia 1110396331 64.698
#> 3 Americas 301139947 78.242
#> 4 Asia 223547000 70.650
#> 5 Americas 190010647 72.390
#> 6 Asia 169270617 65.483
#> 7 Asia 150448339 64.062
#> 8 Africa 135031164 46.859
#> 9 Asia 127467972 82.603
#> 10 Americas 108700891 76.195
# convert row names to a column
top_10_df <- rownames_to_column(top_10_df, var = "country")
top_10_df
#> country continent population lifeExpectancy
#> 1 China Asia 1318683096 72.961
#> 2 India Asia 1110396331 64.698
#> 3 United States Americas 301139947 78.242
#> 4 Indonesia Asia 223547000 70.650
#> 5 Brazil Americas 190010647 72.390
#> 6 Pakistan Asia 169270617 65.483
#> 7 Bangladesh Asia 150448339 64.062
#> 8 Nigeria Africa 135031164 46.859
#> 9 Japan Asia 127467972 82.603
#> 10 Mexico Americas 108700891 76.195
# convert a column to row names
column_to_rownames(top_10_df, var = "country")
#> continent population lifeExpectancy
#> China Asia 1318683096 72.961
#> India Asia 1110396331 64.698
#> United States Americas 301139947 78.242
#> Indonesia Asia 223547000 70.650
#> Brazil Americas 190010647 72.390
#> Pakistan Asia 169270617 65.483
#> Bangladesh Asia 150448339 64.062
#> Nigeria Africa 135031164 46.859
#> Japan Asia 127467972 82.603
#> Mexico Americas 108700891 76.195
# convert row index to a column
rowid_to_column(top_10_df, var = "rank")
#> rank country continent population lifeExpectancy
#> 1 1 China Asia 1318683096 72.961
#> 2 2 India Asia 1110396331 64.698
#> 3 3 United States Americas 301139947 78.242
#> 4 4 Indonesia Asia 223547000 70.650
#> 5 5 Brazil Americas 190010647 72.390
#> 6 6 Pakistan Asia 169270617 65.483
#> 7 7 Bangladesh Asia 150448339 64.062
#> 8 8 Nigeria Africa 135031164 46.859
#> 9 9 Japan Asia 127467972 82.603
#> 10 10 Mexico Americas 108700891 76.1955.2 Manipulating categorical data with forcats
The package forcats comes with a series of functions all beginning with fct_ for working with categorical data. This package is developed and maintained by Hadley Wickham and is part of the tidyverse universe of packages.
Categorical data in R is represented by factors.
# install.packages(forcats)
library(forcats)
library(gapminder)
# loading data
data(gapminder)
# preparing data
gapminder_2007 <- subset(gapminder, year == 2007, -3)
head(gapminder_2007)
#> # A tibble: 6 x 5
#> country continent lifeExp pop gdpPercap
#> <fct> <fct> <dbl> <int> <dbl>
#> 1 Afghanistan Asia 43.8 31889923 975.
#> 2 Albania Europe 76.4 3600523 5937.
#> 3 Algeria Africa 72.3 33333216 6223.
#> 4 Angola Africa 42.7 12420476 4797.
#> 5 Argentina Americas 75.3 40301927 12779.
#> 6 Australia Oceania 81.2 20434176 34435.
sapply(gapminder_2007, class)
#> country continent lifeExp pop gdpPercap
#> "factor" "factor" "numeric" "integer" "numeric"5.2.1 Inspecting factors
5.2.1.1 Get categories
The functions levels() and fct_unique() are used to get levels or categories.
# get levels using base R
levels(gapminder_2007$continent)
#> [1] "Africa" "Americas" "Asia" "Europe" "Oceania"
# get levels using forcats
fct_unique(gapminder_2007$continent)
#> [1] Africa Americas Asia Europe Oceania
#> Levels: Africa Americas Asia Europe Oceania5.2.1.2 Get the number of categories
The functions nlevels() and length(fct_unique()) are used to get the number of categories or levels.
# get the number of categories using base R
nlevels(gapminder_2007$continent)
#> [1] 5
# get the number of categories using forcats
length(fct_unique(gapminder_2007$continent))
#> [1] 55.2.1.3 Count of values by categories
The function table() and fct_count() are used to get count of values by categories with the later returning a tibble.
# count of elements by categories using base R
table(gapminder_2007$continent)
#>
#> Africa Americas Asia Europe Oceania
#> 52 25 33 30 2
# count of elements by categories using forcats
fct_count(gapminder_2007$continent)
#> # A tibble: 5 x 2
#> f n
#> <fct> <int>
#> 1 Africa 52
#> 2 Americas 25
#> 3 Asia 33
#> 4 Europe 30
#> 5 Oceania 25.2.1.4 Reordering levels
# get levels
table(gapminder_2007$continent)
#>
#> Africa Americas Asia Europe Oceania
#> 52 25 33 30 25.2.1.4.1 Manually reordering levels
The function fct_relevel() is used to manually reorder levels.
# manually reorder levels
gapminder_2007$continent <- fct_relevel(gapminder_2007$continent,
c('Asia', 'Africa', 'Americas', 'Europe', 'Oceania'))
table(gapminder_2007$continent)
#>
#> Asia Africa Americas Europe Oceania
#> 33 52 25 30 2
# oceania first
gapminder_2007$continent <- fct_relevel(gapminder_2007$continent, 'Oceania')
table(gapminder_2007$continent)
#>
#> Oceania Asia Africa Americas Europe
#> 2 33 52 25 305.2.1.5 Reordering levels by frequency of occurrence
The function fct_infreq() reorders levels by the number of times they occur in the data with the highest first.
# ordering levels by the frequency they appear in a dataset
gapminder_2007$continent <- fct_infreq(gapminder_2007$continent, ordered = NA)
table(gapminder_2007$continent)
#>
#> Africa Asia Europe Americas Oceania
#> 52 33 30 25 2The argument ordered = TRUE returns an ordered factor.
# unordered factor
class(fct_infreq(gapminder_2007$continent, ordered = NA))
#> [1] "factor"
# ordered factor
class(fct_infreq(gapminder_2007$continent, ordered = TRUE))
#> [1] "ordered" "factor"5.2.2 Reordering levels by their order in data
The function fct_inorder() reorders levels by the order in which they appear in the data set.
# ordering levels by the order in which they appear in a dataset
gapminder_2007$continent <- fct_inorder(gapminder_2007$continent, ordered = NA)
table(gapminder_2007$continent)
#>
#> Asia Europe Africa Americas Oceania
#> 33 30 52 25 25.2.2.1 Reversing the order
The function fct_rev() reverses the order of the levels.
5.2.2.2 Random order
The function fct_shuffle() randomly shuffles levels.
# randomly shuffling level order
gapminder_2007$continent <- fct_shuffle(gapminder_2007$continent)
table(gapminder_2007$continent)
#>
#> Asia Oceania Africa Europe Americas
#> 33 2 52 30 255.2.2.3 Reordering level by another column
The function fct_reorder() reorders levels by another column or vector.
# ordering levels by another column
gapminder_2007$continent <-
fct_reorder(gapminder_2007$continent, gapminder_2007$pop, .fun = sum, .desc = TRUE)
levels(gapminder_2007$continent)
#> [1] "Asia" "Africa" "Americas" "Europe" "Oceania"
# using median
gapminder_2007$continent <-
fct_reorder(gapminder_2007$continent, gapminder_2007$pop, .fun = median, .desc = TRUE)
levels(gapminder_2007$continent)
#> [1] "Asia" "Oceania" "Africa" "Europe" "Americas"
# ascending
gapminder_2007$continent <-
fct_reorder(gapminder_2007$continent, gapminder_2007$pop, .fun = median, .desc = FALSE)
levels(gapminder_2007$continent)
#> [1] "Americas" "Europe" "Africa" "Oceania" "Asia"
# population by continent
(pop_cont <- aggregate(pop ~ continent, gapminder, sum, subset = year == 2007))
#> continent pop
#> 1 Africa 929539692
#> 2 Americas 898871184
#> 3 Asia 3811953827
#> 4 Europe 586098529
#> 5 Oceania 24549947
# plotting a barchart
with(pop_cont, barplot(pop/1e6, names.arg = continent))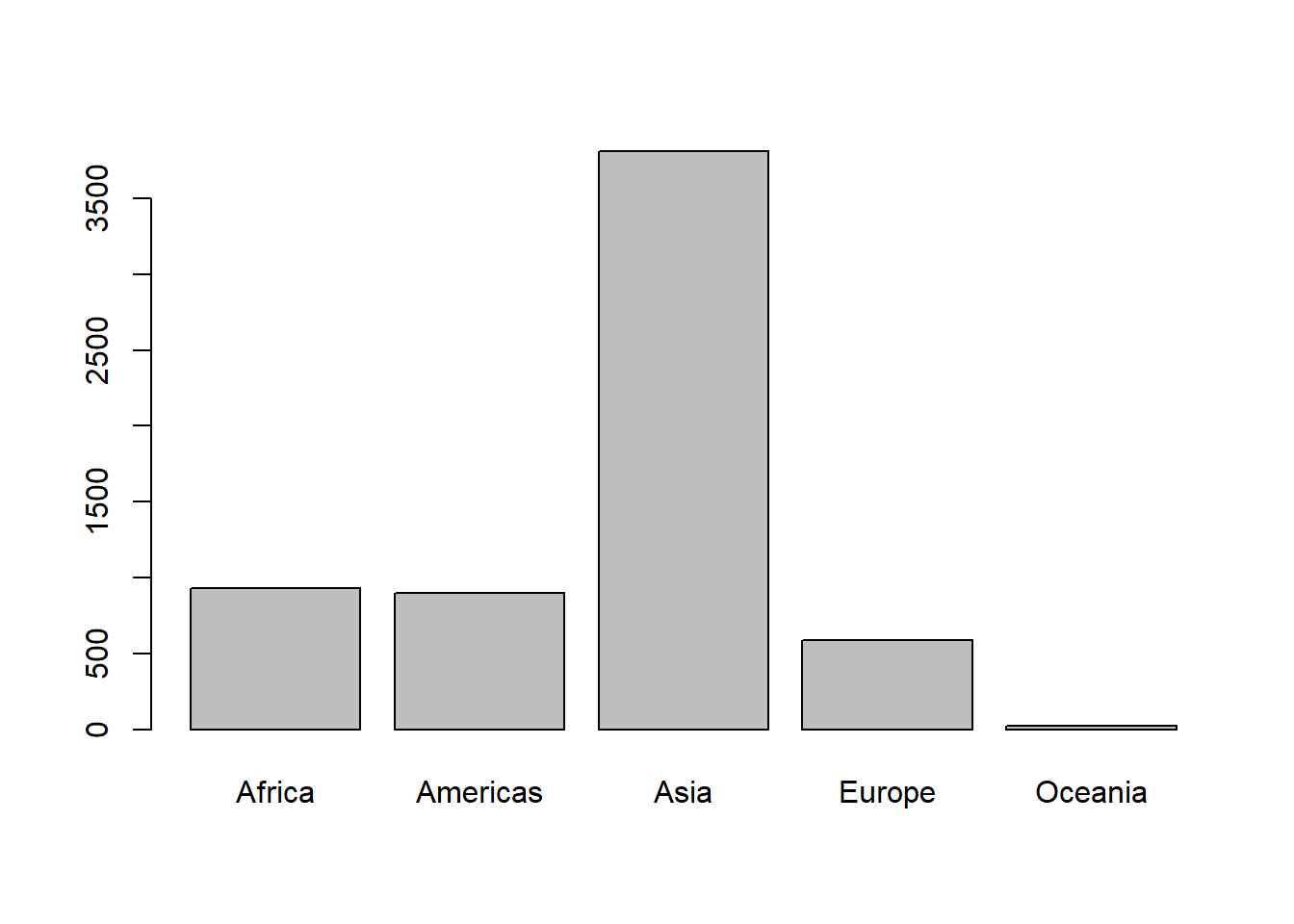
# reordering continent by population
pop_cont$continent <- fct_reorder(pop_cont$continent, pop_cont$pop, .desc = TRUE)
levels(pop_cont$continent)
#> [1] "Asia" "Africa" "Americas" "Europe" "Oceania"
# sorting data frame by continent
pop_cont <- with(pop_cont, pop_cont[order(continent),])
pop_cont
#> continent pop
#> 3 Asia 3811953827
#> 1 Africa 929539692
#> 2 Americas 898871184
#> 4 Europe 586098529
#> 5 Oceania 24549947
# plotting barplot
with(pop_cont, barplot(pop/1e6, names.arg = continent))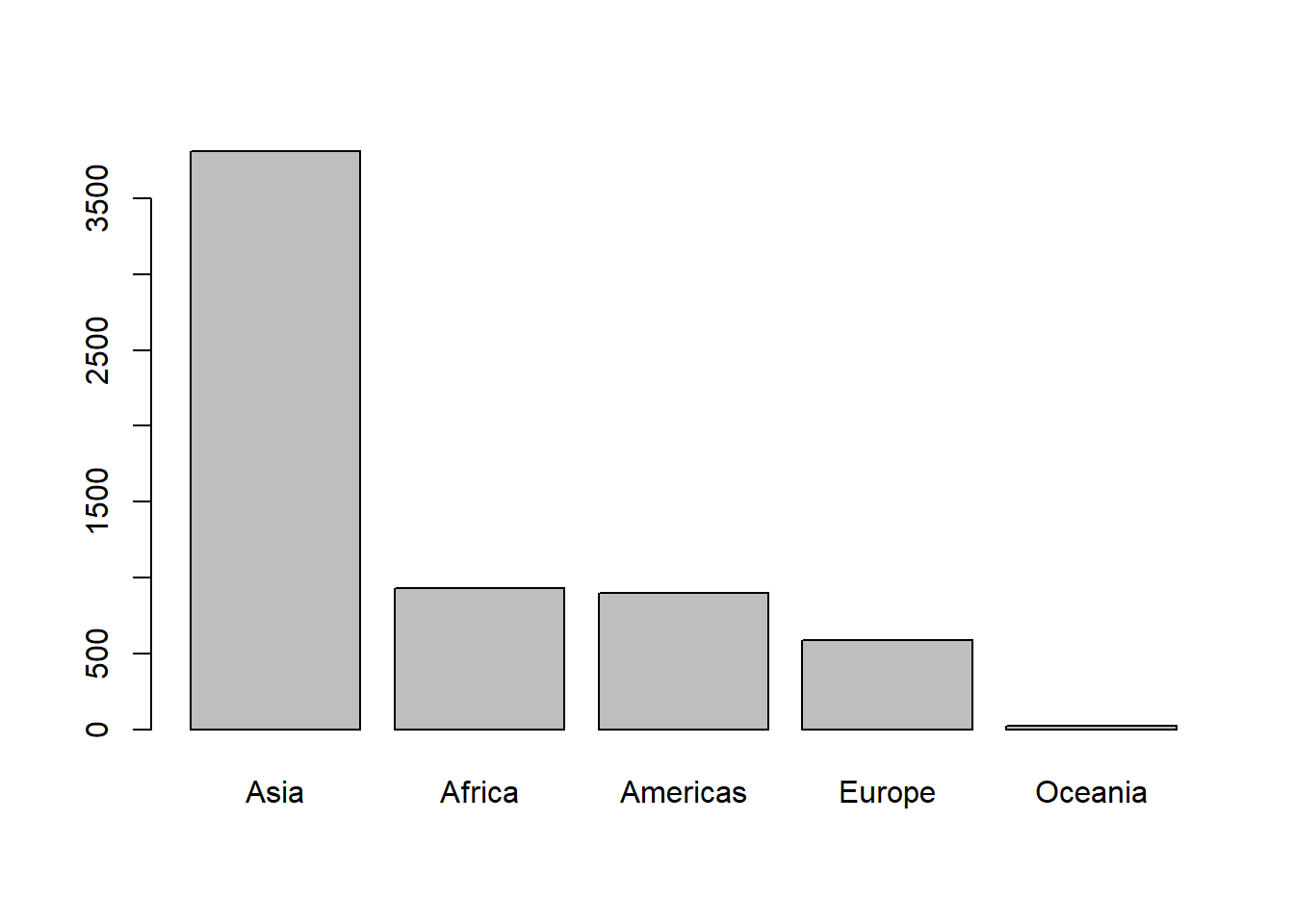
# producing an ascending bar chart
pop_cont$continent <- fct_reorder(pop_cont$continent, pop_cont$pop, .desc = FALSE)
pop_cont <- with(pop_cont, pop_cont[order(continent),])
with(pop_cont, barplot(pop/1e6, names.arg = continent))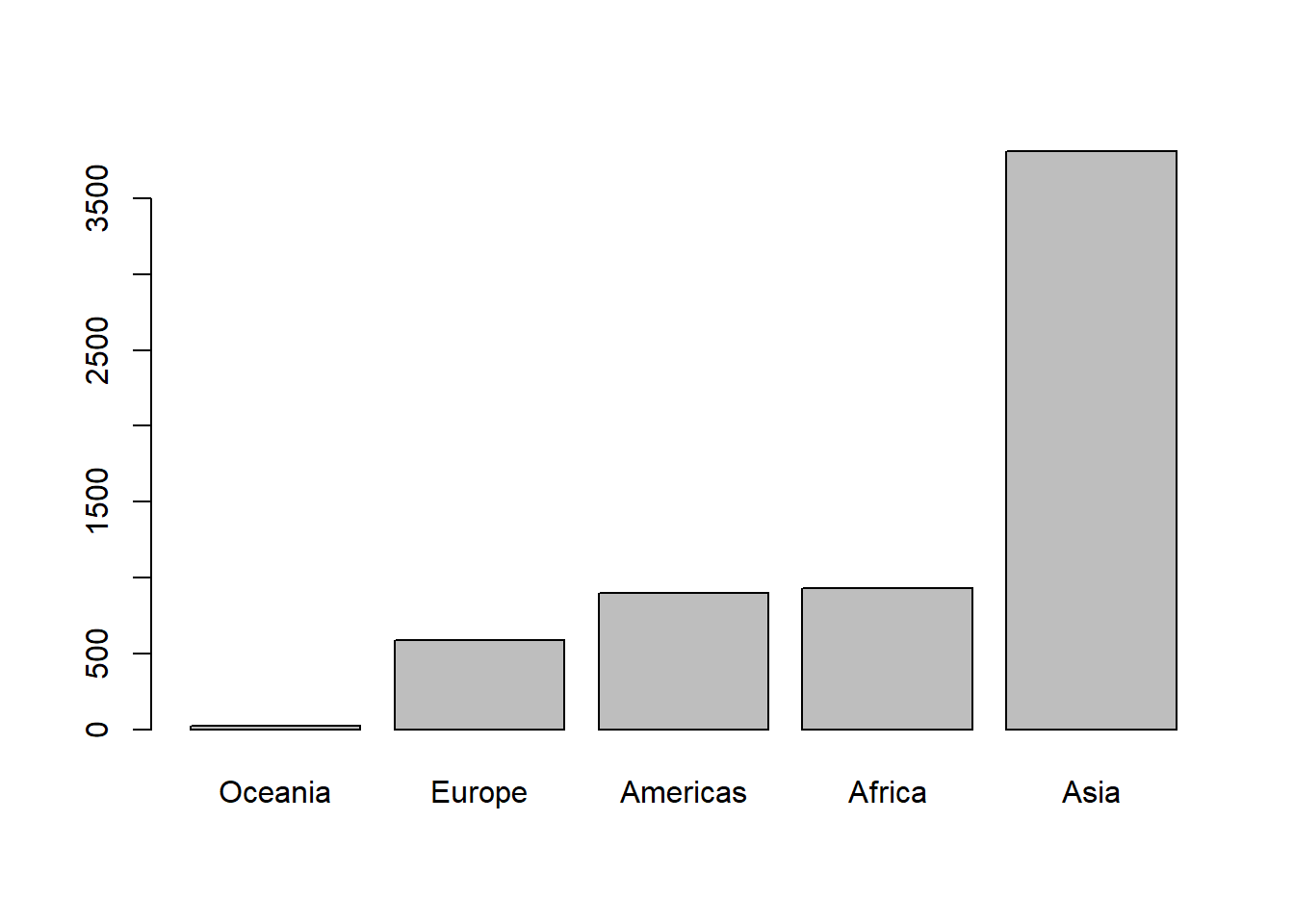
5.2.3 Restructuring levels and their labels
5.2.3.1 Renaming labels
The function fct_recode() is used to rename levels. It takes the form new_name = old_name.
levels(fct_recode(gapminder_2007$continent, 'AS' = 'Asia', 'Af' = 'Africa', 'Eu' = 'Europe'))
#> [1] "Americas" "Eu" "Af" "Oceania" "AS"5.2.3.2 collapsing levels
The function fct_collapse() is used to collapse levels into a new one.
# collapsing europe and africa into euroafrica
gapminder_2007$continent <-
fct_collapse(gapminder_2007$continent, Euroafrica = c('Africa', 'Europe'))
table(gapminder_2007$continent)
#>
#> Americas Euroafrica Oceania Asia
#> 25 82 2 33
# population by continent
(pop_cont <- aggregate(pop ~ continent, gapminder_2007, sum))
#> continent pop
#> 1 Americas 898871184
#> 2 Euroafrica 1515638221
#> 3 Oceania 24549947
#> 4 Asia 38119538275.2.3.3 combining levels
The functions fct_lump() and fct_lump_min() combines levels together based on the frequency of occurrence of each level.
# combining the least frequent levels
gapminder_2007 <- subset(gapminder, year == 2007, -3)
table(fct_lump(gapminder_2007$continent))
#>
#> Africa Asia Europe Other
#> 52 33 30 27Using the arguments n= and p= we can specify the type of combining to perform; with positive values indicating combining rarest levels while negative values indicate combining most common levels.
# combining all except the first most common
table(fct_lump(gapminder_2007$continent, n = 1))
#>
#> Africa Other
#> 52 90
# combining all except the first 2 most common
table(fct_lump(gapminder_2007$continent, n = 2))
#>
#> Africa Asia Other
#> 52 33 57
# combining all except the first 3 most common
table(fct_lump(gapminder_2007$continent, n = 3))
#>
#> Africa Asia Europe Other
#> 52 33 30 27
# combining all except the first rarest
table(fct_lump(gapminder_2007$continent, n = -1))
#>
#> Oceania Other
#> 2 140
# combining all except the first 2 rarest
table(fct_lump(gapminder_2007$continent, n = -2))
#>
#> Americas Oceania Other
#> 25 2 115
# combining all except the first 3 rarest
table(fct_lump(gapminder_2007$continent, n = -3))
#>
#> Americas Europe Oceania Other
#> 25 30 2 85
# using prop positive
table(fct_lump(gapminder_2007$continent, prop = 0.25))
#>
#> Africa Other
#> 52 90
table(fct_lump(gapminder_2007$continent, prop = 0.22))
#>
#> Africa Asia Other
#> 52 33 57
table(fct_lump(gapminder_2007$continent, prop = 0.2))
#>
#> Africa Asia Europe Other
#> 52 33 30 27
# using prop negative
table(fct_lump(gapminder_2007$continent, prop = -0.25))
#>
#> Americas Asia Europe Oceania Other
#> 25 33 30 2 52
table(fct_lump(gapminder_2007$continent, prop = -0.22))
#>
#> Americas Europe Oceania Other
#> 25 30 2 85
table(fct_lump(gapminder_2007$continent, prop = -0.2))
#>
#> Americas Oceania Other
#> 25 2 115With fct_lump_min() combining is done based on whether a threshold declared by the min argument is met.
table(gapminder_2007$continent)
#>
#> Africa Americas Asia Europe Oceania
#> 52 25 33 30 2
# combining levels with less than 25 counts
table(fct_lump_min(gapminder_2007$continent, min = 25))
#>
#> Africa Americas Asia Europe Other
#> 52 25 33 30 2
# combining levels with less than 30 counts
table(fct_lump_min(gapminder_2007$continent, min = 30))
#>
#> Africa Asia Europe Other
#> 52 33 30 27
# combining levels with less than 33 counts
table(fct_lump_min(gapminder_2007$continent, min = 33))
#>
#> Africa Asia Other
#> 52 33 575.2.4 Remove and add levels
5.2.4.1 dropping levels
The function fct_other() will drop levels and replace them with the argument other_level = other by default.
# keeping asia and europe
table(fct_other(gapminder_2007$continent, keep = c('Asia', 'Europe')))
#>
#> Asia Europe Other
#> 33 30 79
# dropping asia and europe
table(fct_other(gapminder_2007$continent, drop = c('Asia', 'Europe')))
#>
#> Africa Americas Oceania Other
#> 52 25 2 63
# replacing other continents with nonEurasia
table(fct_other(gapminder_2007$continent,
keep = c('Asia', 'Europe'),
other_level = 'nonEurasia'))
#>
#> Asia Europe nonEurasia
#> 33 30 79
# replacing europe and asia with Eurasia
table(fct_other(gapminder_2007$continent,
drop = c('Asia', 'Europe'),
other_level = 'Eurasia'))
#>
#> Africa Americas Oceania Eurasia
#> 52 25 2 635.2.5 dropping unused levels
The function fct_drop() is used to drop unused levels. Unused levels are usually a problem while plotting as they appear on the graph though they contain no data.
# dropping Oceania
gapminder_oc <- subset(gapminder_2007, continent != 'Oceania')
table(gapminder_oc$continent)
#>
#> Africa Americas Asia Europe Oceania
#> 52 25 33 30 0
# Because the level Oceania has not been dropped, it appears on the above plot.
plot(gapminder_oc$continent)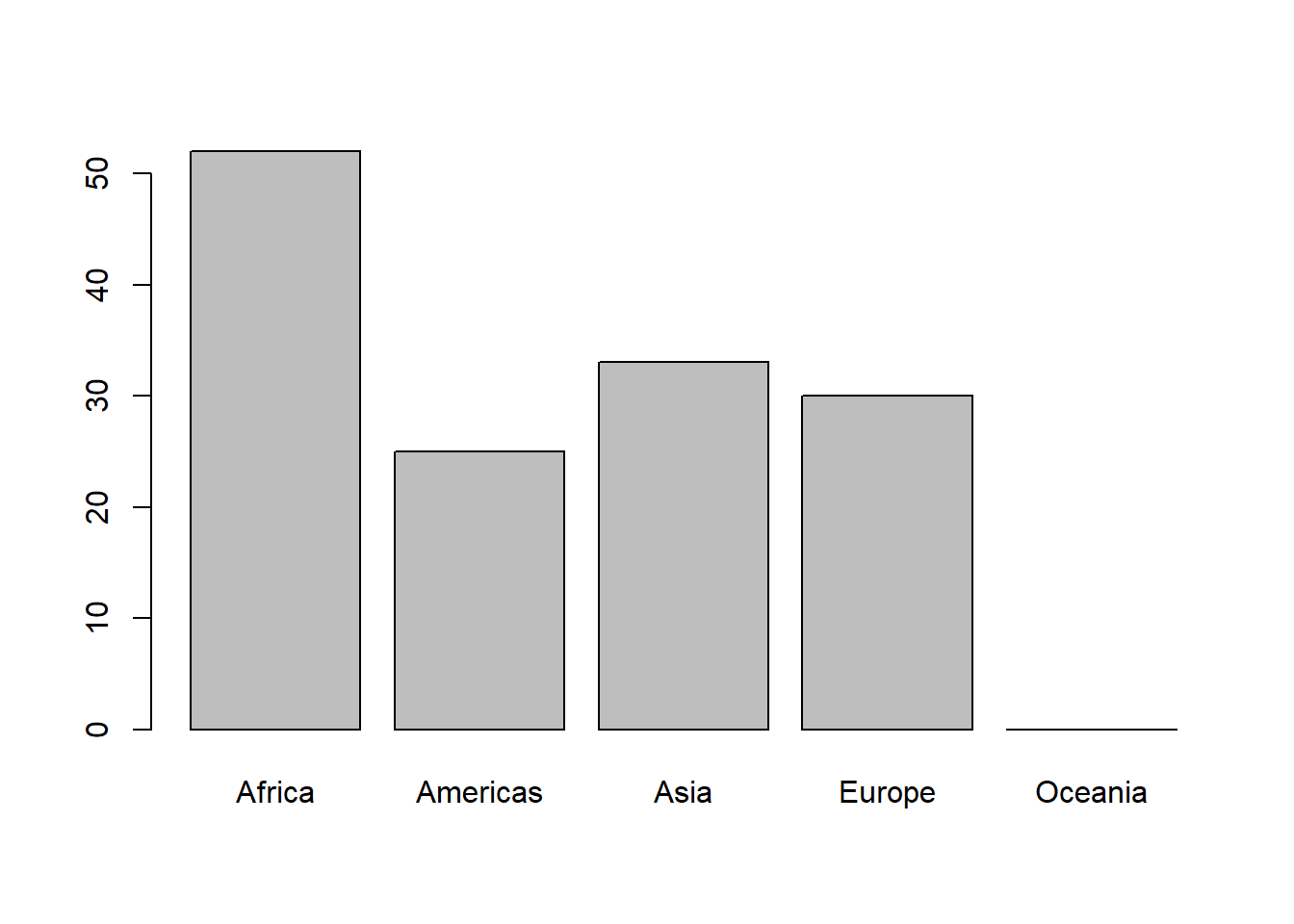
# dropping unused level
table(fct_drop(gapminder_oc$continent))
#>
#> Africa Americas Asia Europe
#> 52 25 33 30
plot(fct_drop(gapminder_oc$continent))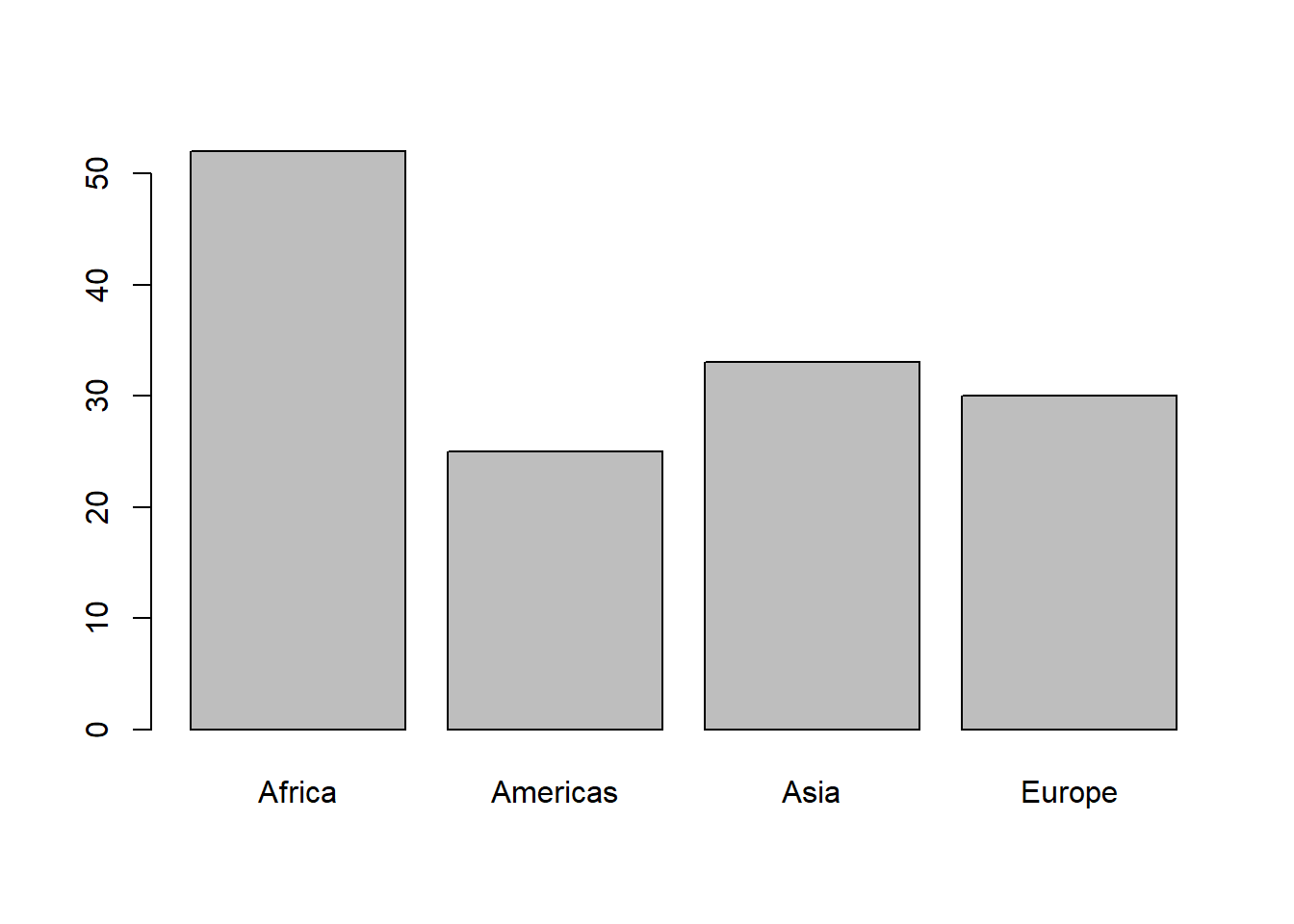
5.2.5.1 adding levels
The function fct_expand() is used to add levels.
# adding the level arctic
table(fct_expand(gapminder_oc$continent, 'arctic'))
#>
#> Africa Americas Asia Europe Oceania arctic
#> 52 25 33 30 0 0
# adding the levels arctic and antarctica
table(fct_expand(gapminder_oc$continent, c('arctic', 'antarctica')))
#>
#> Africa Americas Asia Europe Oceania
#> 52 25 33 30 0
#> arctic antarctica
#> 0 0
# newly added levels appear on the plot though they have no data
plot(fct_expand(gapminder_oc$continent, c('arctic', 'antarctica')))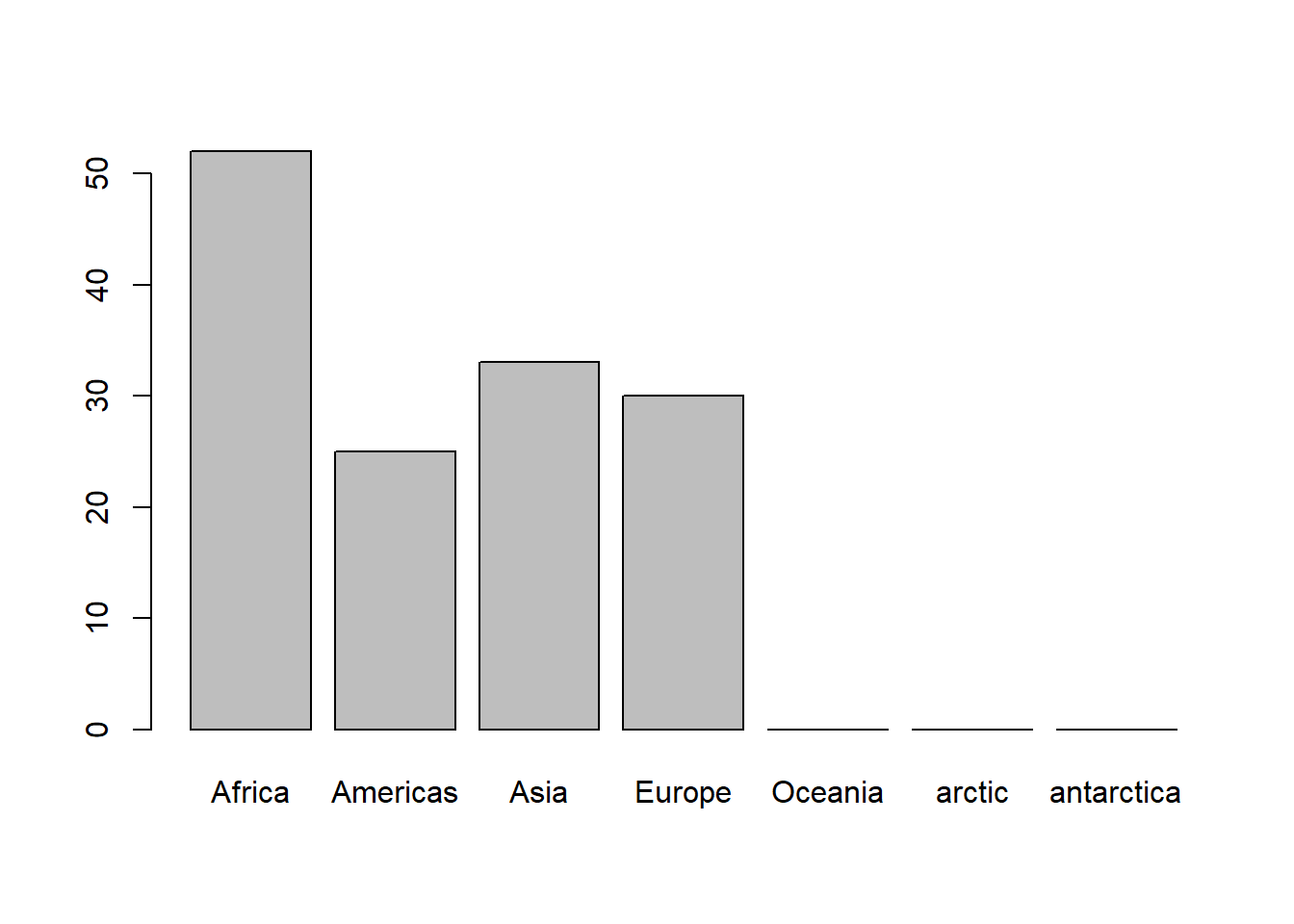
5.3 Data Manipulation with dplyr and tidyr
The package dplyr is one of the core packages in a group of packages known as the tidyverse. It was developed and released in 2014 by Hadley Wickham and others. dplyr is meant to be for data manipulation what ggplot2 is for data visualization, that is the grammar of data manipulation. It focuses solely on data frame manipulation and transformation using a set of verbs (functions) which are consistent and easy to understand.
Since dyplr belongs to the tidyverse world, it can be installed either by installing tidyverse or by installing dplyr itself.
5.3.1 Rename columns and rows
5.3.1.1 Renaming columns
The function rename() is used to rename columns.
library(readr)
library(dplyr)
library(gapminder)
# loading data
data(gapminder)
# get column names
names(gapminder)
#> [1] "country" "continent" "year" "lifeExp"
#> [5] "pop" "gdpPercap"
# set column names
gapminder <- rename(gapminder,
Country = country,
Continent = continent,
Year = year,
`Life Expectancy` = lifeExp,
Population = pop,
`GDP per Capita` = gdpPercap)
# get column names
colnames(gapminder)
#> [1] "Country" "Continent" "Year"
#> [4] "Life Expectancy" "Population" "GDP per Capita"5.3.1.2 Renaming rows
Tibble does not support row names. See this.
5.3.2 Select columns and filter rows
5.3.2.1 Selecting and dropping columns
The function select() is used to select and rename columns.
# preparing data
column_names <- c('Rank', 'Title', 'Genre', 'Description', 'Director', 'Actors',
'Year', 'Runtime', 'Rating', 'Votes', 'Revenue', 'Metascore')
mov <- read.table(file = "data/IMDB-Movie-Data.csv", header = T, sep = ",", dec = ".", fileEncoding = "UTF-8", quote = "\"",
comment.char = "")
head(mov, 3)
#> Rank Title Genre
#> 1 1 Guardians of the Galaxy Action,Adventure,Sci-Fi
#> 2 2 Prometheus Adventure,Mystery,Sci-Fi
#> 3 3 Split Horror,Thriller
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> Year Runtime..Minutes. Rating Votes Revenue..Millions.
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> Metascore
#> 1 76
#> 2 65
#> 3 62
names(mov) <- c('Rank', 'Title', 'Genre', 'Description', 'Director', 'Actors',
'Year', 'Runtime', 'Rating', 'Votes', 'Revenue', 'Metascore')
# selecting columns by column names
movies <- select(mov, c('Title', 'Year', 'Revenue', 'Metascore'))
head(movies, 3)
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
# columns can be passed directly without quotation marks
movies <- select(mov, Title, Year, Revenue, Metascore)
head(movies, 3)
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
# renaming column
movies <- select(mov,
Title,
`Release Year` = Year,
`Revenue in Millions` = Revenue,
Metascore)
head(movies, 3)
#> Title Release Year Revenue in Millions
#> 1 Guardians of the Galaxy 2014 333.13
#> 2 Prometheus 2012 126.46
#> 3 Split 2016 138.12
#> Metascore
#> 1 76
#> 2 65
#> 3 62
# selecting columns by position
movies <- select(mov, 2, 7, 11, 12)
head(movies, 3)
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
# selecting columns by sequencing
movies <- select(mov, 7:12)
head(movies, 3)
#> Year Runtime Rating Votes Revenue Metascore
#> 1 2014 121 8.1 757074 333.13 76
#> 2 2012 124 7.0 485820 126.46 65
#> 3 2016 117 7.3 157606 138.12 62
# : works with column names
movies <- select(mov, Year:Metascore)
head(movies, 3)
#> Year Runtime Rating Votes Revenue Metascore
#> 1 2014 121 8.1 757074 333.13 76
#> 2 2012 124 7.0 485820 126.46 65
#> 3 2016 117 7.3 157606 138.12 62
# dropping columns by column names
movies <- select(mov, -Rank, -Genre, -Description,
-Director, -Actors, -Runtime, -Rating, -Votes)
head(movies, 3)
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
# dropping columns by sequence
movies <- select(mov, -(1:6))
head(movies, 3)
#> Year Runtime Rating Votes Revenue Metascore
#> 1 2014 121 8.1 757074 333.13 76
#> 2 2012 124 7.0 485820 126.46 65
#> 3 2016 117 7.3 157606 138.12 62
movies <- select(mov, -(Rank:Actors))
head(movies, 3)
#> Year Runtime Rating Votes Revenue Metascore
#> 1 2014 121 8.1 757074 333.13 76
#> 2 2012 124 7.0 485820 126.46 65
#> 3 2016 117 7.3 157606 138.12 62
# dropping columns by index position
movies <- select(mov, -c(1, 3, 4, 5, 6, 8, 9, 10))
head(movies, 3)
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
# dropping columns by index position
movies <- select(mov, -1, -3, -4, -5, -6, -8, -9, -10)
head(movies, 3)
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 625.3.2.2 Selecting column based on a condition
The functions starts_with(), ends_with(), matches(), and contains() are used to select columns based on a specific pattern. The function
-
starts_with(): returns columns that start with a specific prefix -
ends_with(): returns columns that end with a specific suffix -
matches(): returns columns that match a particular regex pattern -
contains(): returns columns that contain a particular string
# selecting columns starting with R
movies <- select(mov, starts_with('R'))
head(movies, 3)
#> Rank Runtime Rating Revenue
#> 1 1 121 8.1 333.13
#> 2 2 124 7.0 126.46
#> 3 3 117 7.3 138.12
# selecting columns starting with R and D
movies <- select(mov, starts_with(c('R', 'M')))
head(movies)
#> Rank Runtime Rating Revenue Metascore
#> 1 1 121 8.1 333.13 76
#> 2 2 124 7.0 126.46 65
#> 3 3 117 7.3 138.12 62
#> 4 4 108 7.2 270.32 59
#> 5 5 123 6.2 325.02 40
#> 6 6 103 6.1 45.13 42
# selecting columns containing ea
movies <- select(mov, contains('ea'))
head(movies)
#> Year
#> 1 2014
#> 2 2012
#> 3 2016
#> 4 2016
#> 5 2016
#> 6 2016
# selecting columns ending with r
movies <- select(mov, ends_with('r'))
head(movies)
#> Director Year
#> 1 James Gunn 2014
#> 2 Ridley Scott 2012
#> 3 M. Night Shyamalan 2016
#> 4 Christophe Lourdelet 2016
#> 5 David Ayer 2016
#> 6 Yimou Zhang 2016
# selecting columns ending with r and e
movies <- select(mov, ends_with(c('k','r')))
head(movies)
#> Rank Director Year
#> 1 1 James Gunn 2014
#> 2 2 Ridley Scott 2012
#> 3 3 M. Night Shyamalan 2016
#> 4 4 Christophe Lourdelet 2016
#> 5 5 David Ayer 2016
#> 6 6 Yimou Zhang 20165.3.3 Selecting a single column
Selecting a single column with select() returns a one-column data frame. Often, a vector is wanted instead, to that end there is the function pull().
The function pull() is used to select a single column and return a vector.
movies <- select(mov, c('Title', 'Year', 'Revenue', 'Metascore'))
# using select returns a tibble
head(select(movies, 'Title'), 3)
#> Title
#> 1 Guardians of the Galaxy
#> 2 Prometheus
#> 3 Split
class(select(movies, 'Title'))
#> [1] "data.frame"
# using pull returns a vector whose type depends on the data type of the column
head(pull(movies, var = 1))
#> [1] "Guardians of the Galaxy" "Prometheus"
#> [3] "Split" "Sing"
#> [5] "Suicide Squad" "The Great Wall"
class(pull(movies, var = 1))
#> [1] "character"5.3.4 Filtering rows
The function filter() is used to filter rows.
movies <- select(mov, -c(1, 3, 4, 6, 8, 10))
# using the filter() function
movies. <- filter(movies, Year == 2006)
head(movies., 3)
#> Title
#> 1 The Prestige
#> 2 Pirates of the Caribbean: Dead Man's Chest
#> 3 The Departed
#> Director Year Rating Revenue Metascore
#> 1 Christopher Nolan 2006 8.5 53.08 66
#> 2 Gore Verbinski 2006 7.3 423.03 53
#> 3 Martin Scorsese 2006 8.5 132.37 85
tail(movies., 3)
#> Title
#> 42 Talladega Nights: The Ballad of Ricky Bobby
#> 43 Lucky Number Slevin
#> 44 Inland Empire
#> Director Year Rating Revenue Metascore
#> 42 Adam McKay 2006 6.6 148.21 66
#> 43 Paul McGuigan 2006 7.8 22.49 53
#> 44 David Lynch 2006 7.0 NA NA
# selecting movies released in 2006 with a rating above 8
filter(movies, Year == 2006 & Rating >= 8)
#> Title Director
#> 1 The Prestige Christopher Nolan
#> 2 The Departed Martin Scorsese
#> 3 Casino Royale Martin Campbell
#> 4 Pan's Labyrinth Guillermo del Toro
#> 5 The Lives of Others Florian Henckel von Donnersmarck
#> 6 The Pursuit of Happyness Gabriele Muccino
#> 7 Blood Diamond Edward Zwick
#> Year Rating Revenue Metascore
#> 1 2006 8.5 53.08 66
#> 2 2006 8.5 132.37 85
#> 3 2006 8.0 167.01 80
#> 4 2006 8.2 37.62 98
#> 5 2006 8.5 11.28 89
#> 6 2006 8.0 162.59 64
#> 7 2006 8.0 57.37 64
# without the & operator
filter(movies, Year == 2006, Rating >= 8)
#> Title Director
#> 1 The Prestige Christopher Nolan
#> 2 The Departed Martin Scorsese
#> 3 Casino Royale Martin Campbell
#> 4 Pan's Labyrinth Guillermo del Toro
#> 5 The Lives of Others Florian Henckel von Donnersmarck
#> 6 The Pursuit of Happyness Gabriele Muccino
#> 7 Blood Diamond Edward Zwick
#> Year Rating Revenue Metascore
#> 1 2006 8.5 53.08 66
#> 2 2006 8.5 132.37 85
#> 3 2006 8.0 167.01 80
#> 4 2006 8.2 37.62 98
#> 5 2006 8.5 11.28 89
#> 6 2006 8.0 162.59 64
#> 7 2006 8.0 57.37 64
# selecting rows with NA values on the Metascore column
movies. <- filter(movies, is.na(Metascore))
head(movies.)
#> Title Director Year Rating
#> 1 Paris pieds nus Dominique Abel 2016 6.8
#> 2 Bahubali: The Beginning S.S. Rajamouli 2015 8.3
#> 3 Dead Awake Phillip Guzman 2016 4.7
#> 4 5- 25- 77 Patrick Read Johnson 2007 7.1
#> 5 Don't Fuck in the Woods Shawn Burkett 2016 2.7
#> 6 Fallen Scott Hicks 2016 5.6
#> Revenue Metascore
#> 1 NA NA
#> 2 6.50 NA
#> 3 0.01 NA
#> 4 NA NA
#> 5 NA NA
#> 6 NA NA
# selecting rows with NA values on the Revenue and Metascore column
movies. <- filter(movies, is.na(Revenue), is.na(Metascore))
head(movies.)
#> Title Director Year Rating
#> 1 Paris pieds nus Dominique Abel 2016 6.8
#> 2 5- 25- 77 Patrick Read Johnson 2007 7.1
#> 3 Don't Fuck in the Woods Shawn Burkett 2016 2.7
#> 4 Fallen Scott Hicks 2016 5.6
#> 5 Contratiempo Oriol Paulo 2016 7.9
#> 6 Boyka: Undisputed IV Todor Chapkanov 2016 7.4
#> Revenue Metascore
#> 1 NA NA
#> 2 NA NA
#> 3 NA NA
#> 4 NA NA
#> 5 NA NA
#> 6 NA NA
# selecting rows with NA values on either the Revenue or Metascore column
movies. <- filter(movies, is.na(Revenue) | is.na(Metascore))
head(movies.)
#> Title Director Year Rating
#> 1 Mindhorn Sean Foley 2016 6.4
#> 2 Hounds of Love Ben Young 2016 6.7
#> 3 Paris pieds nus Dominique Abel 2016 6.8
#> 4 Bahubali: The Beginning S.S. Rajamouli 2015 8.3
#> 5 Dead Awake Phillip Guzman 2016 4.7
#> 6 5- 25- 77 Patrick Read Johnson 2007 7.1
#> Revenue Metascore
#> 1 NA 71
#> 2 NA 72
#> 3 NA NA
#> 4 6.50 NA
#> 5 0.01 NA
#> 6 NA NA
# selecting rows without NA values on the Metascore column
movies. <- filter(movies, !is.na(Metascore))
head(movies.)
#> Title Director Year Rating
#> 1 Guardians of the Galaxy James Gunn 2014 8.1
#> 2 Prometheus Ridley Scott 2012 7.0
#> 3 Split M. Night Shyamalan 2016 7.3
#> 4 Sing Christophe Lourdelet 2016 7.2
#> 5 Suicide Squad David Ayer 2016 6.2
#> 6 The Great Wall Yimou Zhang 2016 6.1
#> Revenue Metascore
#> 1 333.13 76
#> 2 126.46 65
#> 3 138.12 62
#> 4 270.32 59
#> 5 325.02 40
#> 6 45.13 42
# selecting rows without NA values on the Revenue and Metascore columns
movies. <- filter(movies, !is.na(Revenue), !is.na(Metascore))
head(movies.)
#> Title Director Year Rating
#> 1 Guardians of the Galaxy James Gunn 2014 8.1
#> 2 Prometheus Ridley Scott 2012 7.0
#> 3 Split M. Night Shyamalan 2016 7.3
#> 4 Sing Christophe Lourdelet 2016 7.2
#> 5 Suicide Squad David Ayer 2016 6.2
#> 6 The Great Wall Yimou Zhang 2016 6.1
#> Revenue Metascore
#> 1 333.13 76
#> 2 126.46 65
#> 3 138.12 62
#> 4 270.32 59
#> 5 325.02 40
#> 6 45.13 42
nrow(movies.)
#> [1] 838
# selecting rows without NA values on either the Revenue or Metascore columns
movies. <- filter(movies, !is.na(Revenue) | !is.na(Metascore))
head(movies.)
#> Title Director Year Rating
#> 1 Guardians of the Galaxy James Gunn 2014 8.1
#> 2 Prometheus Ridley Scott 2012 7.0
#> 3 Split M. Night Shyamalan 2016 7.3
#> 4 Sing Christophe Lourdelet 2016 7.2
#> 5 Suicide Squad David Ayer 2016 6.2
#> 6 The Great Wall Yimou Zhang 2016 6.1
#> Revenue Metascore
#> 1 333.13 76
#> 2 126.46 65
#> 3 138.12 62
#> 4 270.32 59
#> 5 325.02 40
#> 6 45.13 42
nrow(movies.)
#> [1] 970
# selecting films released in 2006 and 2008
movies. <- filter(movies, Year %in% c(2006, 2008))
head(movies.)
#> Title
#> 1 The Dark Knight
#> 2 The Prestige
#> 3 Pirates of the Caribbean: Dead Man's Chest
#> 4 The Departed
#> 5 300
#> 6 Mamma Mia!
#> Director Year Rating Revenue Metascore
#> 1 Christopher Nolan 2008 9.0 533.32 82
#> 2 Christopher Nolan 2006 8.5 53.08 66
#> 3 Gore Verbinski 2006 7.3 423.03 53
#> 4 Martin Scorsese 2006 8.5 132.37 85
#> 5 Zack Snyder 2006 7.7 210.59 52
#> 6 Phyllida Lloyd 2008 6.4 143.70 51
# selecting films released by 'James Gunn' or 'James Marsh'
movies. <- filter(movies, Director %in% c('James Gunn', 'James Marsh'))
head(movies.)
#> Title Director Year Rating Revenue
#> 1 Guardians of the Galaxy James Gunn 2014 8.1 333.13
#> 2 The Theory of Everything James Marsh 2014 7.7 35.89
#> 3 Slither James Gunn 2006 6.5 7.77
#> 4 Super James Gunn 2010 6.8 0.32
#> Metascore
#> 1 76
#> 2 72
#> 3 69
#> 4 50
# selecting films released between 2006 and 2008
movies. <- filter(movies, between(Year, 2006, 2008))
head(movies., 3)
#> Title Director Year Rating Revenue
#> 1 5- 25- 77 Patrick Read Johnson 2007 7.1 NA
#> 2 The Dark Knight Christopher Nolan 2008 9.0 533.32
#> 3 The Prestige Christopher Nolan 2006 8.5 53.08
#> Metascore
#> 1 NA
#> 2 82
#> 3 66
tail(movies., 3)
#> Title Director Year Rating Revenue
#> 147 Taare Zameen Par Aamir Khan 2007 8.5 1.20
#> 148 Hostel: Part II Eli Roth 2007 5.5 17.54
#> 149 Step Up 2: The Streets Jon M. Chu 2008 6.2 58.01
#> Metascore
#> 147 42
#> 148 46
#> 149 505.3.4.1 Randomly selecting rows
The function sample_frac() randomly samples rows and returns a fixed fraction of them.
# sampling by a proportion
sample_frac(movies, 0.005, replace = TRUE)
#> Title Director Year Rating Revenue
#> 1 Bad Moms Jon Lucas 2016 6.2 113.08
#> 2 Rescue Dawn Werner Herzog 2006 7.3 5.48
#> 3 Rescue Dawn Werner Herzog 2006 7.3 5.48
#> 4 The Warriors Gate Matthias Hoene 2016 5.3 NA
#> 5 Marie Antoinette Sofia Coppola 2006 6.4 15.96
#> Metascore
#> 1 60
#> 2 77
#> 3 77
#> 4 77
#> 5 65The function sample_n() randomly samples rows and returns a fixed number of them.
# sampling by number
sample_n(movies, 5, replace = TRUE)
#> Title Director Year Rating Revenue
#> 1 Youth Paolo Sorrentino 2015 7.3 2.70
#> 2 Brimstone Martin Koolhoven 2016 7.1 NA
#> 3 Perfetti sconosciuti Paolo Genovese 2016 7.7 NA
#> 4 Watchmen Zack Snyder 2009 7.6 107.50
#> 5 Footloose Craig Brewer 2011 5.9 51.78
#> Metascore
#> 1 64
#> 2 44
#> 3 43
#> 4 56
#> 5 585.3.5 Slicing
The function slice() is used to slice a data set.
slice(movies, 200:205)
#> Title Director Year Rating
#> 1 Central Intelligence Rawson Marshall Thurber 2016 6.3
#> 2 Edge of Tomorrow Doug Liman 2014 7.9
#> 3 A Cure for Wellness Gore Verbinski 2016 6.5
#> 4 Snowden Oliver Stone 2016 7.3
#> 5 Iron Man Jon Favreau 2008 7.9
#> 6 Allegiant Robert Schwentke 2016 5.7
#> Revenue Metascore
#> 1 127.38 52
#> 2 100.19 71
#> 3 8.10 47
#> 4 21.48 58
#> 5 318.30 79
#> 6 66.00 335.3.6 Top values
The function top_n() returns the top nth number of elements in a column.
# top 5 movies by revenue
top_n(movies, 5, Revenue)
#> Title
#> 1 Star Wars: Episode VII - The Force Awakens
#> 2 The Dark Knight
#> 3 The Avengers
#> 4 Jurassic World
#> 5 Avatar
#> Director Year Rating Revenue Metascore
#> 1 J.J. Abrams 2015 8.1 936.63 81
#> 2 Christopher Nolan 2008 9.0 533.32 82
#> 3 Joss Whedon 2012 8.1 623.28 69
#> 4 Colin Trevorrow 2015 7.0 652.18 59
#> 5 James Cameron 2009 7.8 760.51 83
# if no column is specified, the last is used.
top_n(movies, 5)
#> Title Director Year Rating
#> 1 Manchester by the Sea Kenneth Lonergan 2016 7.9
#> 2 Moonlight Barry Jenkins 2016 7.5
#> 3 12 Years a Slave Steve McQueen 2013 8.1
#> 4 Pan's Labyrinth Guillermo del Toro 2006 8.2
#> 5 Ratatouille Brad Bird 2007 8.0
#> 6 Gravity Alfonso Cuarón 2013 7.8
#> 7 Boyhood Richard Linklater 2014 7.9
#> Revenue Metascore
#> 1 47.70 96
#> 2 27.85 99
#> 3 56.67 96
#> 4 37.62 98
#> 5 206.44 96
#> 6 274.08 96
#> 7 25.36 100The function top_frac() returns the top nth elements in a column by proportion.
# top 0.5% of movies by revenue
top_frac(movies, 0.005, Revenue)
#> Title
#> 1 Star Wars: Episode VII - The Force Awakens
#> 2 The Dark Knight
#> 3 The Avengers
#> 4 Jurassic World
#> 5 Avatar
#> Director Year Rating Revenue Metascore
#> 1 J.J. Abrams 2015 8.1 936.63 81
#> 2 Christopher Nolan 2008 9.0 533.32 82
#> 3 Joss Whedon 2012 8.1 623.28 69
#> 4 Colin Trevorrow 2015 7.0 652.18 59
#> 5 James Cameron 2009 7.8 760.51 83
# if no column is specified, the last is used.
top_frac(movies, 0.005)
#> Title Director Year Rating
#> 1 Manchester by the Sea Kenneth Lonergan 2016 7.9
#> 2 Moonlight Barry Jenkins 2016 7.5
#> 3 12 Years a Slave Steve McQueen 2013 8.1
#> 4 Pan's Labyrinth Guillermo del Toro 2006 8.2
#> 5 Ratatouille Brad Bird 2007 8.0
#> 6 Gravity Alfonso Cuarón 2013 7.8
#> 7 Boyhood Richard Linklater 2014 7.9
#> Revenue Metascore
#> 1 47.70 96
#> 2 27.85 99
#> 3 56.67 96
#> 4 37.62 98
#> 5 206.44 96
#> 6 274.08 96
#> 7 25.36 1005.3.7 Using select and filter
select(filter(mov, Year == 2006, Rating >= 8), 2, 7, 9, 11, 12)
#> Title Year Rating Revenue Metascore
#> 1 The Prestige 2006 8.5 53.08 66
#> 2 The Departed 2006 8.5 132.37 85
#> 3 Casino Royale 2006 8.0 167.01 80
#> 4 Pan's Labyrinth 2006 8.2 37.62 98
#> 5 The Lives of Others 2006 8.5 11.28 89
#> 6 The Pursuit of Happyness 2006 8.0 162.59 64
#> 7 Blood Diamond 2006 8.0 57.37 64
filter(select(mov, 2, 7, 9, 11, 12), Year == 2006, Rating >= 8)
#> Title Year Rating Revenue Metascore
#> 1 The Prestige 2006 8.5 53.08 66
#> 2 The Departed 2006 8.5 132.37 85
#> 3 Casino Royale 2006 8.0 167.01 80
#> 4 Pan's Labyrinth 2006 8.2 37.62 98
#> 5 The Lives of Others 2006 8.5 11.28 89
#> 6 The Pursuit of Happyness 2006 8.0 162.59 64
#> 7 Blood Diamond 2006 8.0 57.37 64With such an operation, it is better to use the pipe operator.
5.3.8 Pipe operator
The pipe operator (%>%) passes an object forward into a function. The shortcut Ctrl + Shift + M for PC and Cmd + Shift + M for Mac is used to insert this operator. Below, we pass the dataset mov into the function filter(), which after processing, passes its output to select().
# passing movies dataset into filter and then to select
mov %>%
filter(Year == 2006 & Rating >= 8) %>%
select(2, 7, 9, 11, 12)
#> Title Year Rating Revenue Metascore
#> 1 The Prestige 2006 8.5 53.08 66
#> 2 The Departed 2006 8.5 132.37 85
#> 3 Casino Royale 2006 8.0 167.01 80
#> 4 Pan's Labyrinth 2006 8.2 37.62 98
#> 5 The Lives of Others 2006 8.5 11.28 89
#> 6 The Pursuit of Happyness 2006 8.0 162.59 64
#> 7 Blood Diamond 2006 8.0 57.37 64
mov %>%
select(2, 7, 9, 11, 12) %>%
filter(Year == 2006 & Rating >= 8)
#> Title Year Rating Revenue Metascore
#> 1 The Prestige 2006 8.5 53.08 66
#> 2 The Departed 2006 8.5 132.37 85
#> 3 Casino Royale 2006 8.0 167.01 80
#> 4 Pan's Labyrinth 2006 8.2 37.62 98
#> 5 The Lives of Others 2006 8.5 11.28 89
#> 6 The Pursuit of Happyness 2006 8.0 162.59 64
#> 7 Blood Diamond 2006 8.0 57.37 64Using . as a placeholder for the data set. The period will be replaced in the function by the data frame or tibble.
mov %>%
filter(.$Year == 2006 & .$Rating >= 8) %>%
select(2, 7, 9, 11, 12)
#> Title Year Rating Revenue Metascore
#> 1 The Prestige 2006 8.5 53.08 66
#> 2 The Departed 2006 8.5 132.37 85
#> 3 Casino Royale 2006 8.0 167.01 80
#> 4 Pan's Labyrinth 2006 8.2 37.62 98
#> 5 The Lives of Others 2006 8.5 11.28 89
#> 6 The Pursuit of Happyness 2006 8.0 162.59 64
#> 7 Blood Diamond 2006 8.0 57.37 645.4 Manipulating Columns
5.4.1 Inserting a new column
The function mutate() and transmutate are used to manipulate columns. They are used to:
- insert new columns
- duplicate columns
- deriving new columns
- update existing ones
# adding a new column known as example
select(mov, c('Title', 'Year', 'Revenue', 'Metascore')) %>%
mutate(example = sample(1000)) %>%
head()
#> Title Year Revenue Metascore example
#> 1 Guardians of the Galaxy 2014 333.13 76 459
#> 2 Prometheus 2012 126.46 65 4
#> 3 Split 2016 138.12 62 130
#> 4 Sing 2016 270.32 59 820
#> 5 Suicide Squad 2016 325.02 40 275
#> 6 The Great Wall 2016 45.13 42 153
# duplicating the Revenue column
select(mov, c('Title', 'Year', 'Revenue', 'Metascore')) %>%
mutate(Metascore.2 = Metascore) %>%
head()
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
#> 4 Sing 2016 270.32 59
#> 5 Suicide Squad 2016 325.02 40
#> 6 The Great Wall 2016 45.13 42
#> Metascore.2
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 42
# deriving the new column Movie Class
labels <- c('Very Low', 'Low', 'Moderate', 'High', 'Very High')
select(mov, c('Title', 'Year', 'Rating', 'Revenue', 'Metascore')) %>%
mutate(`Movie Class` = cut(Rating, breaks = c(0, 5.5, 6.5, 7, 7.5, 10),
labels = labels)) %>%
head()
#> Title Year Rating Revenue Metascore
#> 1 Guardians of the Galaxy 2014 8.1 333.13 76
#> 2 Prometheus 2012 7.0 126.46 65
#> 3 Split 2016 7.3 138.12 62
#> 4 Sing 2016 7.2 270.32 59
#> 5 Suicide Squad 2016 6.2 325.02 40
#> 6 The Great Wall 2016 6.1 45.13 42
#> Movie Class
#> 1 Very High
#> 2 Moderate
#> 3 High
#> 4 High
#> 5 Low
#> 6 Low
# Updating the Director column to uppercase
select(mov, c(Title, Director, Year, Rating, Revenue, Metascore)) %>%
mutate(Director = toupper(Director)) %>%
head()
#> Title Director Year Rating
#> 1 Guardians of the Galaxy JAMES GUNN 2014 8.1
#> 2 Prometheus RIDLEY SCOTT 2012 7.0
#> 3 Split M. NIGHT SHYAMALAN 2016 7.3
#> 4 Sing CHRISTOPHE LOURDELET 2016 7.2
#> 5 Suicide Squad DAVID AYER 2016 6.2
#> 6 The Great Wall YIMOU ZHANG 2016 6.1
#> Revenue Metascore
#> 1 333.13 76
#> 2 126.46 65
#> 3 138.12 62
#> 4 270.32 59
#> 5 325.02 40
#> 6 45.13 42
# using a customized function
# defining a function
fin_crisis <- function(x) {
if(x < 2008){
return('pre financial crisis')
}else if(x < 2010 ){
return('financial crisis')
}else{
return('post financial crisis')
}
}
select(mov, 2, 7, 11, 12) %>%
mutate('fin crisis Class' = sapply(Year, fin_crisis)) %>%
head()
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
#> 4 Sing 2016 270.32 59
#> 5 Suicide Squad 2016 325.02 40
#> 6 The Great Wall 2016 45.13 42
#> fin crisis Class
#> 1 post financial crisis
#> 2 post financial crisis
#> 3 post financial crisis
#> 4 post financial crisis
#> 5 post financial crisis
#> 6 post financial crisis
# deriving a new column from a calculation
select(mov, 2, 5, 7, 8, 11) %>%
mutate('Rev/Run' = round(Revenue/Runtime, 2)) %>%
head()
#> Title Director Year Runtime
#> 1 Guardians of the Galaxy James Gunn 2014 121
#> 2 Prometheus Ridley Scott 2012 124
#> 3 Split M. Night Shyamalan 2016 117
#> 4 Sing Christophe Lourdelet 2016 108
#> 5 Suicide Squad David Ayer 2016 123
#> 6 The Great Wall Yimou Zhang 2016 103
#> Revenue Rev/Run
#> 1 333.13 2.75
#> 2 126.46 1.02
#> 3 138.12 1.18
#> 4 270.32 2.50
#> 5 325.02 2.64
#> 6 45.13 0.44The function case_when() is a condensed form of if else statement or CASE THEN in SQL.
# classifying movies by ratings
select(mov, 2, 7, 9, 11, 12) %>%
mutate(category = case_when(Rating < 5.5 ~ 'Very Low',
Rating < 6.5 ~ 'Low',
Rating < 7 ~ 'Moderate',
Rating < 7.5 ~ 'High',
Rating <= 10 ~ 'Very High')) %>%
head()
#> Title Year Rating Revenue Metascore
#> 1 Guardians of the Galaxy 2014 8.1 333.13 76
#> 2 Prometheus 2012 7.0 126.46 65
#> 3 Split 2016 7.3 138.12 62
#> 4 Sing 2016 7.2 270.32 59
#> 5 Suicide Squad 2016 6.2 325.02 40
#> 6 The Great Wall 2016 6.1 45.13 42
#> category
#> 1 Very High
#> 2 High
#> 3 High
#> 4 High
#> 5 Low
#> 6 LowThe function coalesce() which is modelled after the COALESCE function in SQL returns the first non-missing element. Using it, we can replace NA values in a column.
# selecting some rows containing NA values
select(mov, 2, 5, 7, 9, 11, 12) %>%
filter(is.na(Revenue)) %>%
slice(c(8, 23, 26, 40, 43, 48))
#> Title Director Year Rating
#> 1 The Autopsy of Jane Doe André Ovredal 2016 6.8
#> 2 Old Boy Spike Lee 2013 5.8
#> 3 Satanic Jeffrey G. Hunt 2016 3.7
#> 4 Absolutely Anything Terry Jones 2015 6.0
#> 5 The Headhunter's Calling Mark Williams 2016 6.9
#> 6 Predestination Michael Spierig 2014 7.5
#> Revenue Metascore
#> 1 NA 65
#> 2 NA 49
#> 3 NA NA
#> 4 NA 31
#> 5 NA 85
#> 6 NA 69
# replacing NA values with a value
select(mov, 2, 5, 7, 9, 11, 12) %>%
mutate(Revenue = coalesce(Revenue, 50)) %>%
slice(c(8, 23, 26, 40, 43, 48))
#> Title Director Year Rating
#> 1 Mindhorn Sean Foley 2016 6.4
#> 2 Hounds of Love Ben Young 2016 6.7
#> 3 Paris pieds nus Dominique Abel 2016 6.8
#> 4 5- 25- 77 Patrick Read Johnson 2007 7.1
#> 5 Don't Fuck in the Woods Shawn Burkett 2016 2.7
#> 6 Fallen Scott Hicks 2016 5.6
#> Revenue Metascore
#> 1 50 71
#> 2 50 72
#> 3 50 NA
#> 4 50 NA
#> 5 50 NA
#> 6 50 NA
# replacing NA values with a computed value (mean/median)
select(mov, 2, 5, 7, 9, 11, 12) %>%
mutate(Revenue = coalesce(Revenue, round(median(Revenue, na.rm = T))),
Metascore = coalesce(Metascore, round(mean(Metascore, na.rm = T)))) %>%
slice(c(8, 23, 26, 40, 43, 48))
#> Title Director Year Rating
#> 1 Mindhorn Sean Foley 2016 6.4
#> 2 Hounds of Love Ben Young 2016 6.7
#> 3 Paris pieds nus Dominique Abel 2016 6.8
#> 4 5- 25- 77 Patrick Read Johnson 2007 7.1
#> 5 Don't Fuck in the Woods Shawn Burkett 2016 2.7
#> 6 Fallen Scott Hicks 2016 5.6
#> Revenue Metascore
#> 1 48 71
#> 2 48 72
#> 3 48 59
#> 4 48 59
#> 5 48 59
#> 6 48 59The function transmutate() behaves like mutate() but drops other columns that are not selected.
# transmutate drops unselected columns
select(mov, c(Title, Director, Year, Rating, Revenue, Metascore)) %>%
transmute(Director = toupper(Director)) %>%
head()
#> Director
#> 1 JAMES GUNN
#> 2 RIDLEY SCOTT
#> 3 M. NIGHT SHYAMALAN
#> 4 CHRISTOPHE LOURDELET
#> 5 DAVID AYER
#> 6 YIMOU ZHANG
# transmutate keeps selected columns
select(mov, c(Title, Director, Year, Runtime, Revenue, Metascore)) %>%
transmute(Director = toupper(Director),
Year,
Revenue = round(Revenue/Runtime, 2)) %>%
head()
#> Director Year Revenue
#> 1 JAMES GUNN 2014 2.75
#> 2 RIDLEY SCOTT 2012 1.02
#> 3 M. NIGHT SHYAMALAN 2016 1.18
#> 4 CHRISTOPHE LOURDELET 2016 2.50
#> 5 DAVID AYER 2016 2.64
#> 6 YIMOU ZHANG 2016 0.445.5 Sorting and ranking
5.5.1 Sorting
The function arrange() is used to sort data frames. It does an ascending sort but to do a descending sort, we use the function desc() or the negative sign.
# sort increasing
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
arrange(Revenue) %>%
head(10)
#> Title Year Runtime Revenue Metascore
#> 1 A Kind of Murder 2016 95 0.00 50
#> 2 Dead Awake 2016 99 0.01 NA
#> 3 Wakefield 2016 106 0.01 61
#> 4 Lovesong 2016 84 0.01 74
#> 5 Love, Rosie 2014 102 0.01 44
#> 6 Into the Forest 2015 101 0.01 59
#> 7 Stake Land 2010 98 0.02 66
#> 8 The First Time 2012 95 0.02 55
#> 9 The Blackcoat's Daughter 2015 93 0.02 68
#> 10 The Sea of Trees 2015 110 0.02 23
# sort decreasing using the negative sign
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
arrange(-Revenue) %>%
head(10)
#> Title Year Runtime
#> 1 Star Wars: Episode VII - The Force Awakens 2015 136
#> 2 Avatar 2009 162
#> 3 Jurassic World 2015 124
#> 4 The Avengers 2012 143
#> 5 The Dark Knight 2008 152
#> 6 Rogue One 2016 133
#> 7 Finding Dory 2016 97
#> 8 Avengers: Age of Ultron 2015 141
#> 9 The Dark Knight Rises 2012 164
#> 10 The Hunger Games: Catching Fire 2013 146
#> Revenue Metascore
#> 1 936.63 81
#> 2 760.51 83
#> 3 652.18 59
#> 4 623.28 69
#> 5 533.32 82
#> 6 532.17 65
#> 7 486.29 77
#> 8 458.99 66
#> 9 448.13 78
#> 10 424.65 76
# sort decreasing using desc()
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
arrange(desc(Revenue)) %>%
head(10)
#> Title Year Runtime
#> 1 Star Wars: Episode VII - The Force Awakens 2015 136
#> 2 Avatar 2009 162
#> 3 Jurassic World 2015 124
#> 4 The Avengers 2012 143
#> 5 The Dark Knight 2008 152
#> 6 Rogue One 2016 133
#> 7 Finding Dory 2016 97
#> 8 Avengers: Age of Ultron 2015 141
#> 9 The Dark Knight Rises 2012 164
#> 10 The Hunger Games: Catching Fire 2013 146
#> Revenue Metascore
#> 1 936.63 81
#> 2 760.51 83
#> 3 652.18 59
#> 4 623.28 69
#> 5 533.32 82
#> 6 532.17 65
#> 7 486.29 77
#> 8 458.99 66
#> 9 448.13 78
#> 10 424.65 76
# sorting on multiple columns
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
arrange(-Metascore, Revenue) %>%
head(10)
#> Title Year Runtime Revenue Metascore
#> 1 Boyhood 2014 165 25.36 100
#> 2 Moonlight 2016 111 27.85 99
#> 3 Pan's Labyrinth 2006 118 37.62 98
#> 4 Manchester by the Sea 2016 137 47.70 96
#> 5 12 Years a Slave 2013 134 56.67 96
#> 6 Ratatouille 2007 111 206.44 96
#> 7 Gravity 2013 91 274.08 96
#> 8 Carol 2015 118 0.25 95
#> 9 Zero Dark Thirty 2012 157 95.72 95
#> 10 The Social Network 2010 120 96.92 955.5.2 Ranking
The functions row_number(), ntile(), min_rank(), dense_rank(), percent_rank() and cume_dist() are used for ranking.
# ranking by revenue ascending
select(mov, Title, Year, Revenue, Metascore) %>%
mutate(rank_by_revenue = dense_rank(Revenue)) %>%
head()
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
#> 4 Sing 2016 270.32 59
#> 5 Suicide Squad 2016 325.02 40
#> 6 The Great Wall 2016 45.13 42
#> rank_by_revenue
#> 1 783
#> 2 623
#> 3 646
#> 4 761
#> 5 781
#> 6 370
# ranking by revenue decreasing using desc()
select(mov, Title, Year, Revenue, Metascore) %>%
mutate(rank_by_revenue = dense_rank(desc(Revenue))) %>%
head()
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
#> 4 Sing 2016 270.32 59
#> 5 Suicide Squad 2016 325.02 40
#> 6 The Great Wall 2016 45.13 42
#> rank_by_revenue
#> 1 32
#> 2 192
#> 3 169
#> 4 54
#> 5 34
#> 6 445
# ranking by revenue decreasing using negative sign
select(mov, Title, Year, Revenue, Metascore) %>%
mutate(rank_by_revenue = dense_rank(-Revenue)) %>%
head()
#> Title Year Revenue Metascore
#> 1 Guardians of the Galaxy 2014 333.13 76
#> 2 Prometheus 2012 126.46 65
#> 3 Split 2016 138.12 62
#> 4 Sing 2016 270.32 59
#> 5 Suicide Squad 2016 325.02 40
#> 6 The Great Wall 2016 45.13 42
#> rank_by_revenue
#> 1 32
#> 2 192
#> 3 169
#> 4 54
#> 5 34
#> 6 445
# rank and arrange
select(mov, Title, Year, Revenue, Metascore) %>%
mutate(rank_by_revenue = dense_rank(desc(Revenue))) %>%
arrange(desc(Revenue)) %>%
head()
#> Title Year Revenue
#> 1 Star Wars: Episode VII - The Force Awakens 2015 936.63
#> 2 Avatar 2009 760.51
#> 3 Jurassic World 2015 652.18
#> 4 The Avengers 2012 623.28
#> 5 The Dark Knight 2008 533.32
#> 6 Rogue One 2016 532.17
#> Metascore rank_by_revenue
#> 1 81 1
#> 2 83 2
#> 3 59 3
#> 4 69 4
#> 5 82 5
#> 6 65 6
# adding row numbers
select(mov, Title, Year, Revenue, Metascore) %>%
arrange(Year, Revenue) %>%
mutate(`row number` = row_number()) %>%
head()
#> Title Year Revenue Metascore
#> 1 Idiocracy 2006 0.44 66
#> 2 The Host 2006 2.20 85
#> 3 Perfume: The Story of a Murderer 2006 2.21 56
#> 4 The Fall 2006 2.28 64
#> 5 She's the Man 2006 2.34 45
#> 6 Rescue Dawn 2006 5.48 77
#> row number
#> 1 1
#> 2 2
#> 3 3
#> 4 4
#> 5 5
#> 6 6
# dividing data into evenly sized buckets
movies <-
select(mov, Title, Year, Revenue, Metascore) %>%
arrange(Year) %>%
mutate(buckets = ntile(Revenue, 5))
head(movies)
#> Title Year Revenue
#> 1 The Prestige 2006 53.08
#> 2 Pirates of the Caribbean: Dead Man's Chest 2006 423.03
#> 3 The Departed 2006 132.37
#> 4 300 2006 210.59
#> 5 Casino Royale 2006 167.01
#> 6 Cars 2006 244.05
#> Metascore buckets
#> 1 66 3
#> 2 53 5
#> 3 85 4
#> 4 52 5
#> 5 80 5
#> 6 73 5
table(movies$buckets)
#>
#> 1 2 3 4 5
#> 175 175 174 174 174
# calculating mean by buckets
tapply(movies$Metascore, movies$buckets, function(x)round(mean(x, na.rm = T), 1))
#> 1 2 3 4 5
#> 62.4 57.7 54.3 58.5 64.95.6 Splitting and Merging columns
5.6.1 Splitting columns
The function separate() from the package tidyr is used to split columns.
# reading data
busiestAirports <- read.table(file = "data/busiestAirports.csv",
header = T,
sep=",",
dec = ".",
quote = "\"")
busiestAirports <- select(busiestAirports, c('iata_icao' = 5, 'location', 'country'))
head(busiestAirports)
#> iata_icao location country
#> 1 ATL/KATL Atlanta, Georgia United States
#> 2 PEK/ZBAA Chaoyang-Shunyi, Beijing China
#> 3 DXB/OMDB Garhoud, Dubai United Arab Emirates
#> 4 LAX/KLAX Los Angeles, California United States
#> 5 HND/RJTT Ota, Tokyo Japan
#> 6 ORD/KORD Chicago, Illinois United States
# splitting the column iata_icao into iata and icao
busiest_Airports <-
tidyr::separate(busiestAirports, col = 'iata_icao', into = c('iata', 'icao'), sep = '/')
head(busiest_Airports)
#> iata icao location country
#> 1 ATL KATL Atlanta, Georgia United States
#> 2 PEK ZBAA Chaoyang-Shunyi, Beijing China
#> 3 DXB OMDB Garhoud, Dubai United Arab Emirates
#> 4 LAX KLAX Los Angeles, California United States
#> 5 HND RJTT Ota, Tokyo Japan
#> 6 ORD KORD Chicago, Illinois United StatesAlso, we can make use of mutate() and substring() from base R or str_sub() from stringr to split by position.
# using substring
busiestAirports %>%
mutate(iata = substring(iata_icao, 1, 3), icao = substring(iata_icao, 5, 7)) %>%
select(-1) %>%
head()
#> location country iata icao
#> 1 Atlanta, Georgia United States ATL KAT
#> 2 Chaoyang-Shunyi, Beijing China PEK ZBA
#> 3 Garhoud, Dubai United Arab Emirates DXB OMD
#> 4 Los Angeles, California United States LAX KLA
#> 5 Ota, Tokyo Japan HND RJT
#> 6 Chicago, Illinois United States ORD KOR
# using str_sub
busiestAirports %>%
mutate(iata = stringr::str_sub(iata_icao, 1, 3), icao = stringr::str_sub(iata_icao, 5, 7)) %>%
select(-1) %>%
head()
#> location country iata icao
#> 1 Atlanta, Georgia United States ATL KAT
#> 2 Chaoyang-Shunyi, Beijing China PEK ZBA
#> 3 Garhoud, Dubai United Arab Emirates DXB OMD
#> 4 Los Angeles, California United States LAX KLA
#> 5 Ota, Tokyo Japan HND RJT
#> 6 Chicago, Illinois United States ORD KOR5.6.2 Merging columns
The function unite() from the package tidyr is used to merge columns.
# reading data
busiestAirports <- read.table(file = "data/busiestAirports.csv",
header = T,
sep=",",
dec = ".",
quote = "\"")
busiestAirports <- select(busiestAirports, c('iata_icao' = 5, 'location', 'country'))
head(busiestAirports)
#> iata_icao location country
#> 1 ATL/KATL Atlanta, Georgia United States
#> 2 PEK/ZBAA Chaoyang-Shunyi, Beijing China
#> 3 DXB/OMDB Garhoud, Dubai United Arab Emirates
#> 4 LAX/KLAX Los Angeles, California United States
#> 5 HND/RJTT Ota, Tokyo Japan
#> 6 ORD/KORD Chicago, Illinois United States
# merging the columns iata, icao into iata_icao
busiestAirports <-
tidyr::unite(busiestAirports, location, country, col = `location country`, sep = ', ')
head(busiestAirports)
#> iata_icao location country
#> 1 ATL/KATL Atlanta, Georgia, United States
#> 2 PEK/ZBAA Chaoyang-Shunyi, Beijing, China
#> 3 DXB/OMDB Garhoud, Dubai, United Arab Emirates
#> 4 LAX/KLAX Los Angeles, California, United States
#> 5 HND/RJTT Ota, Tokyo, Japan
#> 6 ORD/KORD Chicago, Illinois, United States5.6.3 Rearranging columns
The function relocate() is used to rearrange columns. It was added with dplr 1.0.0.
# rearranging columns
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(Revenue, Metascore) %>%
head(3)
#> Revenue Metascore Title Year Runtime
#> 1 333.13 76 Guardians of the Galaxy 2014 121
#> 2 126.46 65 Prometheus 2012 124
#> 3 138.12 62 Split 2016 117
# placing year after metascore
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(Year, .after = Metascore) %>%
head(3)
#> Title Runtime Revenue Metascore Year
#> 1 Guardians of the Galaxy 121 333.13 76 2014
#> 2 Prometheus 124 126.46 65 2012
#> 3 Split 117 138.12 62 2016
# placing year before title
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(Year, .before = Title) %>%
head(3)
#> Year Title Runtime Revenue Metascore
#> 1 2014 Guardians of the Galaxy 121 333.13 76
#> 2 2012 Prometheus 124 126.46 65
#> 3 2016 Split 117 138.12 62
# placing year at the end
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(Year, .after = last_col()) %>%
head(3)
#> Title Runtime Revenue Metascore Year
#> 1 Guardians of the Galaxy 121 333.13 76 2014
#> 2 Prometheus 124 126.46 65 2012
#> 3 Split 117 138.12 62 2016
# numeric columns last
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(where(is.numeric), .after = where(is.character)) %>%
head(3)
#> Title Year Runtime Revenue Metascore
#> 1 Guardians of the Galaxy 2014 121 333.13 76
#> 2 Prometheus 2012 124 126.46 65
#> 3 Split 2016 117 138.12 62
# numeric columns first
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(where(is.numeric), .before = where(is.character)) %>%
head(3)
#> Year Runtime Revenue Metascore Title
#> 1 2014 121 333.13 76 Guardians of the Galaxy
#> 2 2012 124 126.46 65 Prometheus
#> 3 2016 117 138.12 62 Split
# selecting character columns only
select(mov, c(Title, Year, Runtime, Revenue, Metascore)) %>%
relocate(where(is.character)) %>%
head(3)
#> Title Year Runtime Revenue Metascore
#> 1 Guardians of the Galaxy 2014 121 333.13 76
#> 2 Prometheus 2012 124 126.46 65
#> 3 Split 2016 117 138.12 625.6.4 Deleting columns of data
There is no special function to delete columns but the function select() can be used to select or drop columns.
5.7 Manipulating Rows
5.7.1 Inserting rows
The function add_row() is used to add row(s) to data frames. It adds:
- single row with add_row(dt, column_name = value)
- multiple rows with add_row(dt, column_name = values)
# adding a single row of data
select(mov, c(2, 5, 7, 9, 11, 12)) %>%
add_row(Title = "the big g",
Director = "goro lovic",
Year = 2015,
Rating = 9.9,
Revenue = 1000,
Metascore = 100) %>%
tail()
#> Title Director Year Rating
#> 996 Secret in Their Eyes Billy Ray 2015 6.2
#> 997 Hostel: Part II Eli Roth 2007 5.5
#> 998 Step Up 2: The Streets Jon M. Chu 2008 6.2
#> 999 Search Party Scot Armstrong 2014 5.6
#> 1000 Nine Lives Barry Sonnenfeld 2016 5.3
#> 1001 the big g goro lovic 2015 9.9
#> Revenue Metascore
#> 996 NA 45
#> 997 17.54 46
#> 998 58.01 50
#> 999 NA 22
#> 1000 19.64 11
#> 1001 1000.00 100
# adding multiple rows of data
select(mov, c(2, 5, 7, 9, 11, 12)) %>%
add_row(Title= c("the big g", "everyday", "true colors"),
Director = c("goro lovic", "fk", "tupac"),
Year = c(2015, 2016, 2014),
Rating = c(9.9, 6.6, 8),
Revenue = c(1000, 250, 350),
Metascore = c(100, 60, 40)) %>%
tail()
#> Title Director Year Rating
#> 998 Step Up 2: The Streets Jon M. Chu 2008 6.2
#> 999 Search Party Scot Armstrong 2014 5.6
#> 1000 Nine Lives Barry Sonnenfeld 2016 5.3
#> 1001 the big g goro lovic 2015 9.9
#> 1002 everyday fk 2016 6.6
#> 1003 true colors tupac 2014 8.0
#> Revenue Metascore
#> 998 58.01 50
#> 999 NA 22
#> 1000 19.64 11
#> 1001 1000.00 100
#> 1002 250.00 60
#> 1003 350.00 40Rows can also be added using the function bind_rows() which is an efficient implementation of do.call(rbind, dfs) in base R.
# adding a single row
select(mov, 2, 5, 7, 9, 11, 12) %>%
bind_rows(list(Title = "the big g",
Director = "goro lovic",
Year = 2015,
Rating = 9.9,
Revenue = 1000,
Metascore = 100)) %>%
tail(3)
#> Title Director Year Rating Revenue
#> 999 Search Party Scot Armstrong 2014 5.6 NA
#> 1000 Nine Lives Barry Sonnenfeld 2016 5.3 19.64
#> 1001 the big g goro lovic 2015 9.9 1000.00
#> Metascore
#> 999 22
#> 1000 11
#> 1001 100
# adding multiple rows
select(mov, 2, 5, 7, 9, 11, 12) %>%
bind_rows(list(Rank= c(1, 3, 5),
Title= c("the big g", "everyday", "true colors"),
Director = c("goro lovic", "fk", "tupac"),
Year = c(2015, 2016, 2014),
Rating = c(9.9, 6.6, 8),
Revenue = c(1000, 250, 350),
Metascore = c(100, 60, 40))) %>%
tail()
#> Title Director Year Rating
#> 998 Step Up 2: The Streets Jon M. Chu 2008 6.2
#> 999 Search Party Scot Armstrong 2014 5.6
#> 1000 Nine Lives Barry Sonnenfeld 2016 5.3
#> 1001 the big g goro lovic 2015 9.9
#> 1002 everyday fk 2016 6.6
#> 1003 true colors tupac 2014 8.0
#> Revenue Metascore Rank
#> 998 58.01 50 NA
#> 999 NA 22 NA
#> 1000 19.64 11 NA
#> 1001 1000.00 100 1
#> 1002 250.00 60 3
#> 1003 350.00 40 5The function rows_insert() which is modelled after the SQL INSERT clause is also used to insert rows. It requires a column with unique value to uniquely identify each row. This function works on two tibbles, therefore the rows to be inserted must be converted to a tibble using the function tibble() before use.
It should be noted that this function was added in version 1.0.0 of the package, therefore you should do well to update your package to make use of it.
The function rows_insert() requires the argument by which identifies the unique row.
# creating a tibble
tb <-
tibble(Rank = 1001,
Title = "the big g",
Director = "goro lovic",
Year = 2015,
Rating = 9.9,
Revenue = 1000,
Metascore = 100)
# inserting a single value
select(mov, 1, 2, 5, 7, 9, 11, 12) %>%
rows_insert(tb, by = "Rank") %>%
tail()
#> Rank Title Director Year
#> 996 996 Secret in Their Eyes Billy Ray 2015
#> 997 997 Hostel: Part II Eli Roth 2007
#> 998 998 Step Up 2: The Streets Jon M. Chu 2008
#> 999 999 Search Party Scot Armstrong 2014
#> 1000 1000 Nine Lives Barry Sonnenfeld 2016
#> 1001 1001 the big g goro lovic 2015
#> Rating Revenue Metascore
#> 996 6.2 NA 45
#> 997 5.5 17.54 46
#> 998 6.2 58.01 50
#> 999 5.6 NA 22
#> 1000 5.3 19.64 11
#> 1001 9.9 1000.00 100
# inserting multiple values
tb <-
tibble(Rank= 1001:1003,
Title= c("the big g", "everyday", "true colors"),
Director = c("goro lovic", "fk", "tupac"),
Year = c(2015, 2016, 2014),
Rating = c(9.9, 6.6, 8),
Revenue = c(1000, 250, 350),
Metascore = c(100, 60, 40))
select(mov, 1, 2, 5, 7, 9, 11, 12) %>%
rows_insert(tb, by = "Rank") %>%
tail()
#> Rank Title Director Year
#> 998 998 Step Up 2: The Streets Jon M. Chu 2008
#> 999 999 Search Party Scot Armstrong 2014
#> 1000 1000 Nine Lives Barry Sonnenfeld 2016
#> 1001 1001 the big g goro lovic 2015
#> 1002 1002 everyday fk 2016
#> 1003 1003 true colors tupac 2014
#> Rating Revenue Metascore
#> 998 6.2 58.01 50
#> 999 5.6 NA 22
#> 1000 5.3 19.64 11
#> 1001 9.9 1000.00 100
#> 1002 6.6 250.00 60
#> 1003 8.0 350.00 405.7.2 Updating rows of data
The function rows_update() and rows_upsert() which are modelled after the SQL UPDATE and UPSERT clauses are used to update row values. While the former updates row values, the later updates existing rows and inserts new ones, if not present. They both required a column with unique value to uniquely identify each row. As with rows_insert(), these functions work on two tibbles, therefore the rows to be inserted must be converted to a tibble using the function tibble() before use.
# updating a single row
tb <-
tibble(Rank = 5,
Title = "the big g",
Director = "goro lovic",
Year = 2015,
Rating = 9.9,
Revenue = 1000,
Metascore = 100)
select(mov, 1, 2, 5, 7, 9, 11, 12) %>%
rows_update(tb, by = "Rank") %>%
head()
#> Rank Title Director Year
#> 1 1 Guardians of the Galaxy James Gunn 2014
#> 2 2 Prometheus Ridley Scott 2012
#> 3 3 Split M. Night Shyamalan 2016
#> 4 4 Sing Christophe Lourdelet 2016
#> 5 5 the big g goro lovic 2015
#> 6 6 The Great Wall Yimou Zhang 2016
#> Rating Revenue Metascore
#> 1 8.1 333.13 76
#> 2 7.0 126.46 65
#> 3 7.3 138.12 62
#> 4 7.2 270.32 59
#> 5 9.9 1000.00 100
#> 6 6.1 45.13 42
# updating multiple rows
tb <-
tibble(Rank= c(1, 3, 5),
Title= c("the big g", "everyday", "true colors"),
Director = c("goro lovic", "fk", "tupac"),
Year = c(2015, 2016, 2014),
Rating = c(9.9, 6.6, 8),
Revenue = c(1000, 250, 350),
Metascore = c(100, 60, 40))
select(mov, 1, 2, 5, 7, 9, 11, 12) %>%
rows_update(tb, by = "Rank") %>%
head()
#> Rank Title Director Year Rating
#> 1 1 the big g goro lovic 2015 9.9
#> 2 2 Prometheus Ridley Scott 2012 7.0
#> 3 3 everyday fk 2016 6.6
#> 4 4 Sing Christophe Lourdelet 2016 7.2
#> 5 5 true colors tupac 2014 8.0
#> 6 6 The Great Wall Yimou Zhang 2016 6.1
#> Revenue Metascore
#> 1 1000.00 100
#> 2 126.46 65
#> 3 250.00 60
#> 4 270.32 59
#> 5 350.00 40
#> 6 45.13 42
# update existing rows and insert new ones
tb <-
tibble(Rank= c(2,3, 1001),
Title= c("the big g", "everyday", "true colors"),
Director = c("goro lovic", "fk", "tupac"),
Year = c(2015, 2016, 2014),
Rating = c(9.9, 6.6, 8),
Revenue = c(1000, 250, 350),
Metascore = c(100, 60, 40))
select(mov, 1, 2, 5, 7, 9, 11, 12) %>%
rows_upsert(tb, by = "Rank") %>%
slice(c(1:5, 1001))
#> Rank Title Director Year
#> 1 1 Guardians of the Galaxy James Gunn 2014
#> 2 2 the big g goro lovic 2015
#> 3 3 everyday fk 2016
#> 4 4 Sing Christophe Lourdelet 2016
#> 5 5 Suicide Squad David Ayer 2016
#> 6 1001 true colors tupac 2014
#> Rating Revenue Metascore
#> 1 8.1 333.13 76
#> 2 9.9 1000.00 100
#> 3 6.6 250.00 60
#> 4 7.2 270.32 59
#> 5 6.2 325.02 40
#> 6 8.0 350.00 405.7.3 Updating a single value
To update a single value, we make use of mutate() with either ifelse() from base R or if_else() from dplyr or replace() from base R. The function if_else() is an implementation of ifelse() in dplyr.
mov %>%
select(1, 2, 5, 7, 9, 11, 12) %>%
filter(Director == 'Christopher Nolan')
#> Rank Title Director Year Rating
#> 1 37 Interstellar Christopher Nolan 2014 8.6
#> 2 55 The Dark Knight Christopher Nolan 2008 9.0
#> 3 65 The Prestige Christopher Nolan 2006 8.5
#> 4 81 Inception Christopher Nolan 2010 8.8
#> 5 125 The Dark Knight Rises Christopher Nolan 2012 8.5
#> Revenue Metascore
#> 1 187.99 74
#> 2 533.32 82
#> 3 53.08 66
#> 4 292.57 74
#> 5 448.13 78
# replacing a value using ifelse()
mov %>%
select(1, 2, 5, 7, 9, 11, 12) %>%
mutate(Director = ifelse(Director == 'Christopher Nolan', 'C. Nolan', Director)) %>%
slice(c(37, 55, 65, 81, 125))
#> Rank Title Director Year Rating Revenue
#> 1 37 Interstellar C. Nolan 2014 8.6 187.99
#> 2 55 The Dark Knight C. Nolan 2008 9.0 533.32
#> 3 65 The Prestige C. Nolan 2006 8.5 53.08
#> 4 81 Inception C. Nolan 2010 8.8 292.57
#> 5 125 The Dark Knight Rises C. Nolan 2012 8.5 448.13
#> Metascore
#> 1 74
#> 2 82
#> 3 66
#> 4 74
#> 5 78
# increasing revenues for movies produced by Christopher Nolan by 20%
mov %>%
select(1, 2, 5, 7, 9, 11, 12) %>%
mutate(Revenue = ifelse(Director == 'Christopher Nolan', Revenue * 1.2, Revenue)) %>%
slice(c(37, 55, 65, 81, 125))
#> Rank Title Director Year Rating
#> 1 37 Interstellar Christopher Nolan 2014 8.6
#> 2 55 The Dark Knight Christopher Nolan 2008 9.0
#> 3 65 The Prestige Christopher Nolan 2006 8.5
#> 4 81 Inception Christopher Nolan 2010 8.8
#> 5 125 The Dark Knight Rises Christopher Nolan 2012 8.5
#> Revenue Metascore
#> 1 225.588 74
#> 2 639.984 82
#> 3 63.696 66
#> 4 351.084 74
#> 5 537.756 78
# replacing a value using if_else()
mov %>%
select(1, 2, 5, 7, 9, 11, 12) %>%
mutate(Director = if_else(Director == 'Christopher Nolan', 'C. Nolan', Director)) %>%
slice(c(37, 55, 65, 81, 125))
#> Rank Title Director Year Rating Revenue
#> 1 37 Interstellar C. Nolan 2014 8.6 187.99
#> 2 55 The Dark Knight C. Nolan 2008 9.0 533.32
#> 3 65 The Prestige C. Nolan 2006 8.5 53.08
#> 4 81 Inception C. Nolan 2010 8.8 292.57
#> 5 125 The Dark Knight Rises C. Nolan 2012 8.5 448.13
#> Metascore
#> 1 74
#> 2 82
#> 3 66
#> 4 74
#> 5 78
# replacing a value using replace()
select(mov, 2, 5, 7, 9, 11, 12) %>%
mutate(Director = replace(Director, Director == 'Christopher Nolan', 'C. Nolan')) %>%
slice(c(37, 55, 65, 81, 125))
#> Title Director Year Rating Revenue
#> 1 Interstellar C. Nolan 2014 8.6 187.99
#> 2 The Dark Knight C. Nolan 2008 9.0 533.32
#> 3 The Prestige C. Nolan 2006 8.5 53.08
#> 4 Inception C. Nolan 2010 8.8 292.57
#> 5 The Dark Knight Rises C. Nolan 2012 8.5 448.13
#> Metascore
#> 1 74
#> 2 82
#> 3 66
#> 4 74
#> 5 78
# increasing revenues for movies produced by Christopher Nolan by 20%
select(mov, 2, 5, 7, 9, 11, 12) %>%
mutate(Revenue = replace(Director, Director == 'Christopher Nolan',
Revenue[Director == 'Christopher Nolan'] * 1.2)) %>%
slice(c(37, 55, 65, 81, 125))
#> Title Director Year Rating
#> 1 Interstellar Christopher Nolan 2014 8.6
#> 2 The Dark Knight Christopher Nolan 2008 9.0
#> 3 The Prestige Christopher Nolan 2006 8.5
#> 4 Inception Christopher Nolan 2010 8.8
#> 5 The Dark Knight Rises Christopher Nolan 2012 8.5
#> Revenue Metascore
#> 1 225.588 74
#> 2 639.984 82
#> 3 63.696 66
#> 4 351.084 74
#> 5 537.756 785.7.4 Deleting rows of data
There is no special function to delete rows but the functions filter() and slice() can be used to keep or drop rows.
5.7.4.1 Unique rows
The function distinct() removes duplicate rows.
5.8 Combine data: concatenate, join and merge
5.8.1 Concatenating tibbles
Combining datasets using bind_rows()
The function bind_rows() acts like rbind() in Base R.
top_5 <- tibble(country = c('China', 'India', 'United States', 'Indonesia', 'Brazil'),
continent = c('Asia', 'Asia', 'Americas', 'Asia', 'Americas'),
population = c(1318683096, 1110396331, 301139947, 223547000, 190010647),
lifeExpectancy = c(72.961, 64.698, 78.242, 70.65, 72.39))
top_5
#> # A tibble: 5 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
top_5_10 <- tibble(country = c('Pakistan', 'Bangladesh', 'Nigeria', 'Japan', 'Mexico'),
continent = c('Asia', 'Asia', 'Africa', 'Asia', 'Americas'),
population = c(169270617, 150448339, 135031164, 127467972, 108700891),
lifeExpectancy = c(65.483, 64.062, 46.859, 82.603, 76.195))
top_5_10
#> # A tibble: 5 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 Pakistan Asia 169270617 65.5
#> 2 Bangladesh Asia 150448339 64.1
#> 3 Nigeria Africa 135031164 46.9
#> 4 Japan Asia 127467972 82.6
#> 5 Mexico Americas 108700891 76.2
# binding data frames
bind_rows(top_5, top_5_10)
#> # A tibble: 10 x 4
#> country continent population lifeExpectancy
#> <chr> <chr> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0
#> 2 India Asia 1110396331 64.7
#> 3 United States Americas 301139947 78.2
#> 4 Indonesia Asia 223547000 70.6
#> 5 Brazil Americas 190010647 72.4
#> 6 Pakistan Asia 169270617 65.5
#> 7 Bangladesh Asia 150448339 64.1
#> 8 Nigeria Africa 135031164 46.9
#> 9 Japan Asia 127467972 82.6
#> 10 Mexico Americas 108700891 76.2
5.8.2 Combining datasets using bind_cols()
The function bind_cols() acts like cbind() in Base R.
country <-
tibble(country = c('China', 'India', 'United States', 'Indonesia', 'Brazil'),
continent = c('Asia', 'Asia', 'Americas', 'Asia', 'Americas'))
country
#> # A tibble: 5 x 2
#> country continent
#> <chr> <chr>
#> 1 China Asia
#> 2 India Asia
#> 3 United States Americas
#> 4 Indonesia Asia
#> 5 Brazil Americas
variables <-
tibble(country = c('China', 'India', 'United States', 'Indonesia', 'Brazil'),
population = c(1318683096, 1110396331, 301139947, 223547000, 190010647),
lifeExpectancy = c(72.961, 64.698, 78.242, 70.65, 72.39),
perCapita = c(4959, 2452, 42952, 3541, 9066))
variables
#> # A tibble: 5 x 4
#> country population lifeExpectancy perCapita
#> <chr> <dbl> <dbl> <dbl>
#> 1 China 1318683096 73.0 4959
#> 2 India 1110396331 64.7 2452
#> 3 United States 301139947 78.2 42952
#> 4 Indonesia 223547000 70.6 3541
#> 5 Brazil 190010647 72.4 9066
# binding data frames
bind_cols(country, variables[-1])
#> # A tibble: 5 x 5
#> country continent population lifeExpectancy perCapita
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 China Asia 1318683096 73.0 4959
#> 2 India Asia 1110396331 64.7 2452
#> 3 United States Americas 301139947 78.2 42952
#> 4 Indonesia Asia 223547000 70.6 3541
#> 5 Brazil Americas 190010647 72.4 90665.8.3 Joining data frames using Set Operations
group_one <-
tibble(country = c('Ethiopia', 'Congo, Dem. Rep.', 'Egypt', 'United States',
'Mexico', 'India', 'Pakistan', 'Thailand', 'Japan'),
population = c(76511887, 64606759, 80264543, 301139947, 108700891,
1110396331, 169270617, 65068149, 127467972))
group_one
#> # A tibble: 9 x 2
#> country population
#> <chr> <dbl>
#> 1 Ethiopia 76511887
#> 2 Congo, Dem. Rep. 64606759
#> 3 Egypt 80264543
#> 4 United States 301139947
#> 5 Mexico 108700891
#> 6 India 1110396331
#> 7 Pakistan 169270617
#> 8 Thailand 65068149
#> 9 Japan 127467972
group_two <-
tibble(country = c('Ethiopia', 'Vietnam', 'Bangladesh', 'Thailand', 'India'),
population = c(76511887, 85262356, 150448339, 65068149, 111039633))
group_two
#> # A tibble: 5 x 2
#> country population
#> <chr> <dbl>
#> 1 Ethiopia 76511887
#> 2 Vietnam 85262356
#> 3 Bangladesh 150448339
#> 4 Thailand 65068149
#> 5 India 1110396335.8.3.1 Intersection
The function intersect() keeps rows that appear in both datasets.
intersect(group_one, group_two)
#> # A tibble: 2 x 2
#> country population
#> <chr> <dbl>
#> 1 Ethiopia 76511887
#> 2 Thailand 650681495.8.4 Union
The function union() keeps rows that appear in either of the datasets.
5.8.5 Differences
The function setdiff() keeps rows that appear in the first dataset but not in the second.
setdiff(group_one, group_two)
#> # A tibble: 7 x 2
#> country population
#> <chr> <dbl>
#> 1 Congo, Dem. Rep. 64606759
#> 2 Egypt 80264543
#> 3 United States 301139947
#> 4 Mexico 108700891
#> 5 India 1110396331
#> 6 Pakistan 169270617
#> 7 Japan 1274679725.8.5.1 SQL like joins
# preparing data
employees <- tibble(
name = c('john', 'mary', 'david', 'paul', 'susan', 'cynthia', 'Joss', 'dennis'),
age = c(45, 55, 35, 58, 40, 30, 39, 25),
gender = c('m', 'f', 'm', 'm', 'f', 'f', 'm', 'm'),
salary =c(40000, 50000, 35000, 25000, 48000, 32000, 20000, 45000),
department = c('commercial', 'production', NA, 'human resources',
'commercial', 'commercial', 'production', NA))
employees
#> # A tibble: 8 x 5
#> name age gender salary department
#> <chr> <dbl> <chr> <dbl> <chr>
#> 1 john 45 m 40000 commercial
#> 2 mary 55 f 50000 production
#> 3 david 35 m 35000 <NA>
#> 4 paul 58 m 25000 human resources
#> 5 susan 40 f 48000 commercial
#> 6 cynthia 30 f 32000 commercial
#> 7 Joss 39 m 20000 production
#> 8 dennis 25 m 45000 <NA>
departments <- tibble(
department = c('commercial', 'human resources', 'production', 'finance', 'maintenance'),
location = c('washington', 'london', 'paris', 'dubai', 'dublin'))
departments
#> # A tibble: 5 x 2
#> department location
#> <chr> <chr>
#> 1 commercial washington
#> 2 human resources london
#> 3 production paris
#> 4 finance dubai
#> 5 maintenance dublin5.8.6 Left join
The left join returns all records from the left dataset and the matched records from the right dataset. The result is NULL from the right side if there is no match.
left_join(employees, departments)
#> # A tibble: 8 x 6
#> name age gender salary department location
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial washington
#> 2 mary 55 f 50000 production paris
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources london
#> 5 susan 40 f 48000 commercial washington
#> 6 cynthia 30 f 32000 commercial washington
#> 7 Joss 39 m 20000 production paris
#> 8 dennis 25 m 45000 <NA> <NA>5.8.7 Right join
The right join returns all records from the right dataset, and the matched records from the left dataset. The result is NULL from the left side when there is no match.
right_join(employees, departments)
#> # A tibble: 8 x 6
#> name age gender salary department location
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial washington
#> 2 mary 55 f 50000 production paris
#> 3 paul 58 m 25000 human resources london
#> 4 susan 40 f 48000 commercial washington
#> 5 cynthia 30 f 32000 commercial washington
#> 6 Joss 39 m 20000 production paris
#> 7 <NA> NA <NA> NA finance dubai
#> 8 <NA> NA <NA> NA maintenance dublin
# reversing tables produces the same results as a left join
right_join(departments, employees)
#> # A tibble: 8 x 6
#> department location name age gender salary
#> <chr> <chr> <chr> <dbl> <chr> <dbl>
#> 1 commercial washington john 45 m 40000
#> 2 commercial washington susan 40 f 48000
#> 3 commercial washington cynthia 30 f 32000
#> 4 human resources london paul 58 m 25000
#> 5 production paris mary 55 f 50000
#> 6 production paris Joss 39 m 20000
#> 7 <NA> <NA> david 35 m 35000
#> 8 <NA> <NA> dennis 25 m 450005.8.8 Inner join
The inner join selects records that have matching values in both datasets
inner_join(employees, departments)
#> # A tibble: 6 x 6
#> name age gender salary department location
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial washington
#> 2 mary 55 f 50000 production paris
#> 3 paul 58 m 25000 human resources london
#> 4 susan 40 f 48000 commercial washington
#> 5 cynthia 30 f 32000 commercial washington
#> 6 Joss 39 m 20000 production paris5.8.9 Full join
The full join returns all records between the left and right dataset
full_join(employees, departments)
#> # A tibble: 10 x 6
#> name age gender salary department location
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial washington
#> 2 mary 55 f 50000 production paris
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources london
#> 5 susan 40 f 48000 commercial washington
#> 6 cynthia 30 f 32000 commercial washington
#> 7 Joss 39 m 20000 production paris
#> 8 dennis 25 m 45000 <NA> <NA>
#> 9 <NA> NA <NA> NA finance dubai
#> 10 <NA> NA <NA> NA maintenance dublin5.8.10 Anti join
The anti join returns all records found on the left dataset but absent in the right one
anti_join(employees, departments)
#> # A tibble: 2 x 5
#> name age gender salary department
#> <chr> <dbl> <chr> <dbl> <chr>
#> 1 david 35 m 35000 <NA>
#> 2 dennis 25 m 45000 <NA>Tibbles with different column names
# recreating employee table with different column names
employees <- tibble(
name = c('john', 'mary', 'david', 'paul', 'susan', 'cynthia', 'Joss', 'dennis'),
age = c(45, 55, 35, 58, 40, 30, 39, 25),
gender = c('m', 'f', 'm', 'm', 'f', 'f', 'm', 'm'),
salary =c(40000, 50000, 35000, 25000, 48000, 32000, 20000, 45000),
dep_name = c('commercial', 'production', NA, 'human resources',
'commercial', 'commercial', 'production', NA))
employees
#> # A tibble: 8 x 5
#> name age gender salary dep_name
#> <chr> <dbl> <chr> <dbl> <chr>
#> 1 john 45 m 40000 commercial
#> 2 mary 55 f 50000 production
#> 3 david 35 m 35000 <NA>
#> 4 paul 58 m 25000 human resources
#> 5 susan 40 f 48000 commercial
#> 6 cynthia 30 f 32000 commercial
#> 7 Joss 39 m 20000 production
#> 8 dennis 25 m 45000 <NA>
left_join(employees, departments, by = c('dep_name' = 'department'))
#> # A tibble: 8 x 6
#> name age gender salary dep_name location
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial washington
#> 2 mary 55 f 50000 production paris
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources london
#> 5 susan 40 f 48000 commercial washington
#> 6 cynthia 30 f 32000 commercial washington
#> 7 Joss 39 m 20000 production paris
#> 8 dennis 25 m 45000 <NA> <NA>Joining on more than one joining column
# adding a subdepartment
employees <- tibble(
name = c('john', 'mary', 'david', 'paul', 'susan', 'cynthia', 'Joss', 'dennis'),
age = c(45, 55, 35, 58, 40, 30, 39, 25),
gender = c('m', 'f', 'm', 'm', 'f', 'f', 'm', 'm'),
salary =c(40000, 50000, 35000, 25000, 48000, 32000, 20000, 45000),
department = c('commercial', 'production', NA, 'human resources', 'commercial',
'commercial', 'production', NA),
subdepartment = c('marketing', 'production', NA, 'human resources', 'sales',
'sales', 'production', NA))
employees
#> # A tibble: 8 x 6
#> name age gender salary department subdepartment
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial marketing
#> 2 mary 55 f 50000 production production
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources human resourc~
#> 5 susan 40 f 48000 commercial sales
#> 6 cynthia 30 f 32000 commercial sales
#> 7 Joss 39 m 20000 production production
#> 8 dennis 25 m 45000 <NA> <NA>
departments <- tibble(
department = c('commercial', 'commercial', 'human resources', 'production',
'finance', 'finance', 'maintenance'),
subdepartment = c('marketing', 'sales', 'human resources', 'production', 'finance',
'accounting', 'maintenance'),
location = c('washington', 'washington', 'london', 'paris', 'dubai', 'dubai', 'dublin'))
departments
#> # A tibble: 7 x 3
#> department subdepartment location
#> <chr> <chr> <chr>
#> 1 commercial marketing washington
#> 2 commercial sales washington
#> 3 human resources human resources london
#> 4 production production paris
#> 5 finance finance dubai
#> 6 finance accounting dubai
#> 7 maintenance maintenance dublin
# since columns have the same names, joining is done automatically
left_join(employees, departments)
#> # A tibble: 8 x 7
#> name age gender salary department subdepartment
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial marketing
#> 2 mary 55 f 50000 production production
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources human resourc~
#> 5 susan 40 f 48000 commercial sales
#> 6 cynthia 30 f 32000 commercial sales
#> 7 Joss 39 m 20000 production production
#> 8 dennis 25 m 45000 <NA> <NA>
#> # ... with 1 more variable: location <chr>
# declaring column names explicitly
left_join(employees, departments, by = c("department", "subdepartment"))
#> # A tibble: 8 x 7
#> name age gender salary department subdepartment
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial marketing
#> 2 mary 55 f 50000 production production
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources human resourc~
#> 5 susan 40 f 48000 commercial sales
#> 6 cynthia 30 f 32000 commercial sales
#> 7 Joss 39 m 20000 production production
#> 8 dennis 25 m 45000 <NA> <NA>
#> # ... with 1 more variable: location <chr>
# with different names
employees <- tibble(
name = c('john', 'mary', 'david', 'paul', 'susan', 'cynthia', 'Joss', 'dennis'),
age = c(45, 55, 35, 58, 40, 30, 39, 25),
gender = c('m', 'f', 'm', 'm', 'f', 'f', 'm', 'm'),
salary =c(40000, 50000, 35000, 25000, 48000, 32000, 20000, 45000),
dep = c('commercial', 'production', NA, 'human resources', 'commercial',
'commercial', 'production', NA),
sub = c('marketing', 'production', NA, 'human resources', 'sales',
'sales', 'production', NA))
employees
#> # A tibble: 8 x 6
#> name age gender salary dep sub
#> <chr> <dbl> <chr> <dbl> <chr> <chr>
#> 1 john 45 m 40000 commercial marketing
#> 2 mary 55 f 50000 production production
#> 3 david 35 m 35000 <NA> <NA>
#> 4 paul 58 m 25000 human resources human resourc~
#> 5 susan 40 f 48000 commercial sales
#> 6 cynthia 30 f 32000 commercial sales
#> 7 Joss 39 m 20000 production production
#> 8 dennis 25 m 45000 <NA> <NA>
left_join(employees, departments, by = c('dep' = 'department', 'sub' = 'subdepartment'))
#> # A tibble: 8 x 7
#> name age gender salary dep sub location
#> <chr> <dbl> <chr> <dbl> <chr> <chr> <chr>
#> 1 john 45 m 40000 commercial mark~ washing~
#> 2 mary 55 f 50000 production prod~ paris
#> 3 david 35 m 35000 <NA> <NA> <NA>
#> 4 paul 58 m 25000 human resources huma~ london
#> 5 susan 40 f 48000 commercial sales washing~
#> 6 cynthia 30 f 32000 commercial sales washing~
#> 7 Joss 39 m 20000 production prod~ paris
#> 8 dennis 25 m 45000 <NA> <NA> <NA>5.9 Aggregating and grouping data
5.9.1 Aggregating
The function summarise() aggregates data using various summarization functions from both Base R and dplyr itself. In addition to the summarization functions like mean(), median(), sum(), etc. Which come with base R, dplyr comes with the following:
-
n()for counts of rows, -
n_distinct()for counts of unique elements -
first()for first value -
last()for last value -
nth()for nth value
data(gapminder)
# performing aggregations
gapminder %>%
filter(year == 2007) %>%
summarize(`total pop` = sum(pop, na.rm = T),
`mean pop` = mean(pop, na.rm = T),
`median pop` = median(pop, na.rm = T),
`country count` = n())
#> # A tibble: 1 x 4
#> `total pop` `mean pop` `median pop` `country count`
#> <dbl> <dbl> <dbl> <int>
#> 1 6251013179 44021220. 10517531 142The function summarise_at() affects variables selected with a character vector or vars().
# using multiple summarization function
gapminder %>%
filter(year == 2007) %>%
summarise_at(vars(lifeExp), list(mean = mean, median = median, count = ~n()))
#> # A tibble: 1 x 3
#> mean median count
#> <dbl> <dbl> <int>
#> 1 67.0 71.9 142
gapminder %>%
filter(year == 2007) %>%
summarise_at(vars(lifeExp), list(~ mean(.), ~ median(.), ~ n()))
#> # A tibble: 1 x 3
#> mean median n
#> <dbl> <dbl> <int>
#> 1 67.0 71.9 142
# multiple columns with vars
gapminder %>%
filter(year == 2007) %>%
summarise_at(vars(lifeExp, gdpPercap), list(mean = mean, median = median))
#> # A tibble: 1 x 4
#> lifeExp_mean gdpPercap_mean lifeExp_median
#> <dbl> <dbl> <dbl>
#> 1 67.0 11680. 71.9
#> # ... with 1 more variable: gdpPercap_median <dbl>
# multiple columns with vectors
gapminder %>%
filter(year == 2007) %>%
summarise_at(c('lifeExp', 'gdpPercap'), list(mean = mean, median = median))
#> # A tibble: 1 x 4
#> lifeExp_mean gdpPercap_mean lifeExp_median
#> <dbl> <dbl> <dbl>
#> 1 67.0 11680. 71.9
#> # ... with 1 more variable: gdpPercap_median <dbl>
# using a custom function
gapminder %>%
filter(year == 2007) %>%
summarise_at(vars(lifeExp, gdpPercap), list(mean = function(x)round(mean(x), 1),
median = function(x)round(median(x), 1)))
#> # A tibble: 1 x 4
#> lifeExp_mean gdpPercap_mean lifeExp_median
#> <dbl> <dbl> <dbl>
#> 1 67 11680. 71.9
#> # ... with 1 more variable: gdpPercap_median <dbl>5.9.2 Grouping data
The function group_by() is used to group data while the function ungroup() is used to ungroup data after applying grouping. It is always a good idea to ungroup data after working with groupings as functions in dplyr will behave differently with grouped data.
# grouping by single column (continent)
gapminder %>%
filter(year == 2007) %>%
group_by(continent) %>%
summarize(`total pop` = sum(pop, na.rm = T),
`mean pop` = mean(pop, na.rm = T),
`median pop` = median(pop, na.rm = T),
`country count` = n()) %>%
ungroup()
#> # A tibble: 5 x 5
#> continent `total pop` `mean pop` `median pop`
#> <fct> <dbl> <dbl> <dbl>
#> 1 Africa 929539692 17875763. 10093310.
#> 2 Americas 898871184 35954847. 9319622
#> 3 Asia 3811953827 115513752. 24821286
#> 4 Europe 586098529 19536618. 9493598
#> 5 Oceania 24549947 12274974. 12274974.
#> # ... with 1 more variable: country count <int>
# grouping by two categorical columns (continent and year)
gapminder %>%
filter(year %in% c(1987, 2007)) %>%
group_by(continent, year) %>%
summarize(`total pop` = sum(pop, na.rm = T),
`mean pop` = mean(pop, na.rm = T),
`median pop` = median(pop, na.rm = T),
`country count` = n()) %>%
ungroup()
#> # A tibble: 10 x 6
#> continent year `total pop` `mean pop` `median pop`
#> <fct> <int> <dbl> <dbl> <dbl>
#> 1 Africa 1987 574834110 11054502. 6635612.
#> 2 Africa 2007 929539692 17875763. 10093310.
#> 3 Americas 1987 682753971 27310159. 6655297
#> 4 Americas 2007 898871184 35954847. 9319622
#> 5 Asia 1987 2871220762 87006690. 16495304
#> 6 Asia 2007 3811953827 115513752. 24821286
#> 7 Europe 1987 543094160 18103139. 9101370.
#> 8 Europe 2007 586098529 19536618. 9493598
#> 9 Oceania 1987 19574415 9787208. 9787208.
#> 10 Oceania 2007 24549947 12274974. 12274974.
#> # ... with 1 more variable: country count <int>
# sorting by group
gap_data <-
gapminder %>%
group_by(year) %>%
arrange(pop, .by_group = TRUE) %>%
ungroup()
head(gap_data)
#> # A tibble: 6 x 6
#> country continent year lifeExp pop gdpPercap
#> <fct> <fct> <int> <dbl> <int> <dbl>
#> 1 Sao Tome and Principe Africa 1952 46.5 60011 880.
#> 2 Djibouti Africa 1952 34.8 63149 2670.
#> 3 Bahrain Asia 1952 50.9 120447 9867.
#> 4 Iceland Europe 1952 72.5 147962 7268.
#> 5 Comoros Africa 1952 40.7 153936 1103.
#> 6 Kuwait Asia 1952 55.6 160000 108382.
tail(gap_data)
#> # A tibble: 6 x 6
#> country continent year lifeExp pop gdpPercap
#> <fct> <fct> <int> <dbl> <int> <dbl>
#> 1 Pakistan Asia 2007 65.5 169270617 2606.
#> 2 Brazil Americas 2007 72.4 190010647 9066.
#> 3 Indonesia Asia 2007 70.6 223547000 3541.
#> 4 United States Americas 2007 78.2 301139947 42952.
#> 5 India Asia 2007 64.7 1110396331 2452.
#> 6 China Asia 2007 73.0 1318683096 4959.
# ranking by group
select(mov, Title, Year, Revenue, Metascore) %>%
arrange(Year, Revenue) %>%
group_by(Year) %>%
mutate(rank_by_revenue = rank(Revenue, ties.method = "first")) %>%
ungroup() %>%
slice(43:47)
#> # A tibble: 5 x 5
#> Title Year Revenue Metascore rank_by_revenue
#> <chr> <int> <dbl> <int> <int>
#> 1 Deja Vu 2006 NA NA 43
#> 2 Inland Empire 2006 NA NA 44
#> 3 The Babysitters 2007 0.04 35 1
#> 4 Taare Zameen Par 2007 1.2 42 2
#> 5 Funny Games 2007 1.29 NA 3
## NB: Notice as ranking restarts once as 2007 is reached.5.9.3 Splitting data frame by groups
The group_split() is like base::split() in that it splits a data frame.
movies_year <-
select(mov, Title, Year, Revenue, Metascore) %>%
group_split(Year)
length(movies_year)
#> [1] 11
movies_year[1]
#> <list_of<
#> tbl_df<
#> Title : character
#> Year : integer
#> Revenue : double
#> Metascore: integer
#> >
#> >[1]>
#> [[1]]
#> # A tibble: 44 x 4
#> Title Year Revenue Metascore
#> <chr> <int> <dbl> <int>
#> 1 The Prestige 2006 53.1 66
#> 2 Pirates of the Caribbean: Dead M~ 2006 423. 53
#> 3 The Departed 2006 132. 85
#> 4 300 2006 211. 52
#> 5 Casino Royale 2006 167. 80
#> 6 Cars 2006 244. 73
#> 7 Pan's Labyrinth 2006 37.6 98
#> 8 Apocalypto 2006 50.9 68
#> 9 Children of Men 2006 35.3 84
#> 10 The Devil Wears Prada 2006 125. 62
#> # ... with 34 more rows5.10 Pivoting and unpivoting data with tidyr
5.10.1 Pivoting
The function pivot_wider() pivots data that is converting it from long to wide. It expects the following:
- names_from: rows to move to columns
- values_from: values to be placed between the intersection of rows and columns (cell values)
library(tidyr)
# preparing data
dt <-
gapminder %>%
filter(year %in% c(1987, 1997, 2007)) %>%
group_by(continent, year) %>%
summarize(total_pop = sum(pop, na.rm = T)) %>%
ungroup()
dt
#> # A tibble: 15 x 3
#> continent year total_pop
#> <fct> <int> <dbl>
#> 1 Africa 1987 574834110
#> 2 Africa 1997 743832984
#> 3 Africa 2007 929539692
#> 4 Americas 1987 682753971
#> 5 Americas 1997 796900410
#> 6 Americas 2007 898871184
#> 7 Asia 1987 2871220762
#> 8 Asia 1997 3383285500
#> 9 Asia 2007 3811953827
#> 10 Europe 1987 543094160
#> 11 Europe 1997 568944148
#> 12 Europe 2007 586098529
#> 13 Oceania 1987 19574415
#> 14 Oceania 1997 22241430
#> 15 Oceania 2007 24549947
# pivoting data
dt %>%
pivot_wider(names_from = year, values_from = total_pop, names_prefix = 'Y')
#> # A tibble: 5 x 4
#> continent Y1987 Y1997 Y2007
#> <fct> <dbl> <dbl> <dbl>
#> 1 Africa 574834110 743832984 929539692
#> 2 Americas 682753971 796900410 898871184
#> 3 Asia 2871220762 3383285500 3811953827
#> 4 Europe 543094160 568944148 586098529
#> 5 Oceania 19574415 22241430 245499475.10.2 Unpivoting
The function pivot_longer() unpivots data, that is converting it from wide to long. It expects:
- cols: columns to move to row
- names_to: name of the new column for moved columns
- values_to: name of the new column for moved cell values
# preparing data
dt_wide <-
dt %>%
pivot_wider(names_from = year, values_from = total_pop, names_prefix = 'Y')
dt_wide
#> # A tibble: 5 x 4
#> continent Y1987 Y1997 Y2007
#> <fct> <dbl> <dbl> <dbl>
#> 1 Africa 574834110 743832984 929539692
#> 2 Americas 682753971 796900410 898871184
#> 3 Asia 2871220762 3383285500 3811953827
#> 4 Europe 543094160 568944148 586098529
#> 5 Oceania 19574415 22241430 24549947
# unpivoting data
dt_wide %>%
pivot_longer(cols = c(Y1987, Y1997, Y2007)) %>%
head()
#> # A tibble: 6 x 3
#> continent name value
#> <fct> <chr> <dbl>
#> 1 Africa Y1987 574834110
#> 2 Africa Y1997 743832984
#> 3 Africa Y2007 929539692
#> 4 Americas Y1987 682753971
#> 5 Americas Y1997 796900410
#> 6 Americas Y2007 898871184
# replacing name and value
dt_wide %>%
pivot_longer(cols = c(Y1987, Y1997, Y2007), names_to = 'year', values_to = 'population') %>%
head()
#> # A tibble: 6 x 3
#> continent year population
#> <fct> <chr> <dbl>
#> 1 Africa Y1987 574834110
#> 2 Africa Y1997 743832984
#> 3 Africa Y2007 929539692
#> 4 Americas Y1987 682753971
#> 5 Americas Y1997 796900410
#> 6 Americas Y2007 8988711845.11 Dealing with duplicate values with dplyr
The function distinct() is used to extract unique values while n_distinct() returns the count of unique values.
library(readr)
library(dplyr)
# reading data
movies <- read.table(file = "data/IMDB-Movie-Data.csv", header = T, sep = ",", dec = ".", fileEncoding = "UTF-8", quote = "\"",
comment.char = "")
# preparing data
movies %>%
select(7, 12) %>%
filter(Year == 2006) %>%
arrange(Metascore) %>%
head()
#> Year Metascore
#> 1 2006 36
#> 2 2006 45
#> 3 2006 45
#> 4 2006 45
#> 5 2006 46
#> 6 2006 47
# extracting unique values
movies %>%
select(7, 12) %>%
filter(Year == 2006) %>%
arrange(Metascore) %>%
distinct() %>%
head()
#> Year Metascore
#> 1 2006 36
#> 2 2006 45
#> 3 2006 46
#> 4 2006 47
#> 5 2006 48
#> 6 2006 51
# count of unique values
movies %>%
select(7, 12) %>%
filter(Year == 2006) %>%
arrange(Year, Metascore) %>%
n_distinct()
#> [1] 27
# extracting unique values by column
movies %>%
arrange(Year, Metascore) %>%
distinct(Year)
#> Year
#> 1 2006
#> 2 2007
#> 3 2008
#> 4 2009
#> 5 2010
#> 6 2011
#> 7 2012
#> 8 2013
#> 9 2014
#> 10 2015
#> 11 2016
# keeping other columns
movies %>%
select(7, 12) %>%
arrange(Year, Metascore) %>%
distinct(Year, .keep_all= TRUE)
#> Year Metascore
#> 1 2006 36
#> 2 2007 29
#> 3 2008 15
#> 4 2009 23
#> 5 2010 20
#> 6 2011 31
#> 7 2012 31
#> 8 2013 18
#> 9 2014 22
#> 10 2015 18
#> 11 2016 115.12 Dealing with NA values with tidyr
5.12.1 Replacing missing values by LOCF
The function fill() performs NA replacement both by LOCF and NOCB.
library(tidyr)
# reading data
movies <- read.table(file = "data/IMDB-Movie-Data.csv", header = T, sep = ",", dec = ".", fileEncoding = "UTF-8", quote = "\"",
comment.char = "")
names(movies)[c(2,7,11,12)] <- c('Title', 'Year', 'RevenueMillions', 'Metascore')
# replacing NA values to values that precede it
movies %>%
dplyr::arrange(Year) %>%
fill(RevenueMillions, .direction = "down") %>%
tail(10)
#> Rank Title
#> 991 948 King Cobra
#> 992 950 Kicks
#> 993 965 Custody
#> 994 967 L'odyssée
#> 995 975 Queen of Katwe
#> 996 976 My Big Fat Greek Wedding 2
#> 997 978 Amateur Night
#> 998 979 It's Only the End of the World
#> 999 981 Miracles from Heaven
#> 1000 1000 Nine Lives
#> Genre
#> 991 Crime,Drama
#> 992 Adventure
#> 993 Drama
#> 994 Adventure,Biography
#> 995 Biography,Drama,Sport
#> 996 Comedy,Family,Romance
#> 997 Comedy
#> 998 Drama
#> 999 Biography,Drama,Family
#> 1000 Comedy,Family,Fantasy
#> Description
#> 991 This ripped-from-the-headlines drama covers the early rise of gay porn headliner Sean Paul Lockhart a.k.a. Brent Corrigan, before his falling out with the producer who made him famous. When... See full summary »
#> 992 Brandon is a 15 year old whose dream is a pair of fresh Air Jordans. Soon after he gets his hands on them, they're stolen by a local hood, causing Brandon and his two friends to go on a dangerous mission through Oakland to retrieve them.
#> 993 The lives of three women are unexpectedly changed when they cross paths at a New York Family Court.
#> 994 Highly influential and a fearlessly ambitious pioneer, innovator, filmmaker, researcher and conservationist, Jacques-Yves Cousteau's aquatic adventure covers roughly thirty years of an inarguably rich in achievements life.
#> 995 A Ugandan girl sees her world rapidly change after being introduced to the game of chess.
#> 996 A Portokalos family secret brings the beloved characters back together for an even bigger and Greeker wedding.
#> 997 Guy Carter is an award-winning graduate student of architecture. He's got a beautiful wife and a baby on the way. The problem? He doesn't have "his ducks in a row," which only fuels his ... See full summary »
#> 998 Louis (Gaspard Ulliel), a terminally ill writer, returns home after a long absence to tell his family that he is dying.
#> 999 A young girl suffering from a rare digestive disorder finds herself miraculously cured after surviving a terrible accident.
#> 1000 A stuffy businessman finds himself trapped inside the body of his family's cat.
#> Director
#> 991 Justin Kelly
#> 992 Justin Tipping
#> 993 James Lapine
#> 994 Jérôme Salle
#> 995 Mira Nair
#> 996 Kirk Jones
#> 997 Lisa Addario
#> 998 Xavier Dolan
#> 999 Patricia Riggen
#> 1000 Barry Sonnenfeld
#> Actors
#> 991 Garrett Clayton, Christian Slater, Molly Ringwald,James Kelley
#> 992 Jahking Guillory, Christopher Jordan Wallace,Christopher Meyer, Kofi Siriboe
#> 993 Viola Davis, Hayden Panettiere, Catalina Sandino Moreno, Ellen Burstyn
#> 994 Lambert Wilson, Pierre Niney, Audrey Tautou,Laurent Lucas
#> 995 Madina Nalwanga, David Oyelowo, Lupita Nyong'o, Martin Kabanza
#> 996 Nia Vardalos, John Corbett, Michael Constantine, Lainie Kazan
#> 997 Jason Biggs, Janet Montgomery,Ashley Tisdale, Bria L. Murphy
#> 998 Nathalie Baye, Vincent Cassel, Marion Cotillard, Léa Seydoux
#> 999 Jennifer Garner, Kylie Rogers, Martin Henderson,Brighton Sharbino
#> 1000 Kevin Spacey, Jennifer Garner, Robbie Amell,Cheryl Hines
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 991 2016 91 5.6 3990 0.03
#> 992 2016 80 6.1 2417 0.15
#> 993 2016 104 6.9 280 0.15
#> 994 2016 122 6.7 1810 0.15
#> 995 2016 124 7.4 6753 8.81
#> 996 2016 94 6.0 20966 59.57
#> 997 2016 92 5.0 2229 59.57
#> 998 2016 97 7.0 10658 59.57
#> 999 2016 109 7.0 12048 61.69
#> 1000 2016 87 5.3 12435 19.64
#> Metascore
#> 991 48
#> 992 69
#> 993 72
#> 994 70
#> 995 73
#> 996 37
#> 997 38
#> 998 48
#> 999 44
#> 1000 115.12.2 Replacing missing values by NOCB
# replacing NA values with proceeding values
fill(movies, RevenueMillions, .direction = "up") %>%
head(10)
#> Rank Title Genre
#> 1 1 Guardians of the Galaxy Action,Adventure,Sci-Fi
#> 2 2 Prometheus Adventure,Mystery,Sci-Fi
#> 3 3 Split Horror,Thriller
#> 4 4 Sing Animation,Comedy,Family
#> 5 5 Suicide Squad Action,Adventure,Fantasy
#> 6 6 The Great Wall Action,Adventure,Fantasy
#> 7 7 La La Land Comedy,Drama,Music
#> 8 8 Mindhorn Comedy
#> 9 9 The Lost City of Z Action,Adventure,Biography
#> 10 10 Passengers Adventure,Drama,Romance
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> 4 In a city of humanoid animals, a hustling theater impresario's attempt to save his theater with a singing competition becomes grander than he anticipates even as its finalists' find that their lives will never be the same.
#> 5 A secret government agency recruits some of the most dangerous incarcerated super-villains to form a defensive task force. Their first mission: save the world from the apocalypse.
#> 6 European mercenaries searching for black powder become embroiled in the defense of the Great Wall of China against a horde of monstrous creatures.
#> 7 A jazz pianist falls for an aspiring actress in Los Angeles.
#> 8 A has-been actor best known for playing the title character in the 1980s detective series "Mindhorn" must work with the police when a serial killer says that he will only speak with Detective Mindhorn, whom he believes to be a real person.
#> 9 A true-life drama, centering on British explorer Col. Percival Fawcett, who disappeared while searching for a mysterious city in the Amazon in the 1920s.
#> 10 A spacecraft traveling to a distant colony planet and transporting thousands of people has a malfunction in its sleep chambers. As a result, two passengers are awakened 90 years early.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> 4 Christophe Lourdelet
#> 5 David Ayer
#> 6 Yimou Zhang
#> 7 Damien Chazelle
#> 8 Sean Foley
#> 9 James Gray
#> 10 Morten Tyldum
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> 4 Matthew McConaughey,Reese Witherspoon, Seth MacFarlane, Scarlett Johansson
#> 5 Will Smith, Jared Leto, Margot Robbie, Viola Davis
#> 6 Matt Damon, Tian Jing, Willem Dafoe, Andy Lau
#> 7 Ryan Gosling, Emma Stone, Rosemarie DeWitt, J.K. Simmons
#> 8 Essie Davis, Andrea Riseborough, Julian Barratt,Kenneth Branagh
#> 9 Charlie Hunnam, Robert Pattinson, Sienna Miller, Tom Holland
#> 10 Jennifer Lawrence, Chris Pratt, Michael Sheen,Laurence Fishburne
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> 4 2016 108 7.2 60545 270.32
#> 5 2016 123 6.2 393727 325.02
#> 6 2016 103 6.1 56036 45.13
#> 7 2016 128 8.3 258682 151.06
#> 8 2016 89 6.4 2490 8.01
#> 9 2016 141 7.1 7188 8.01
#> 10 2016 116 7.0 192177 100.01
#> Metascore
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 42
#> 7 93
#> 8 71
#> 9 78
#> 10 41
# on more than one column
fill(movies, c(RevenueMillions, Metascore), .direction = "up") %>%
head(10)
#> Rank Title Genre
#> 1 1 Guardians of the Galaxy Action,Adventure,Sci-Fi
#> 2 2 Prometheus Adventure,Mystery,Sci-Fi
#> 3 3 Split Horror,Thriller
#> 4 4 Sing Animation,Comedy,Family
#> 5 5 Suicide Squad Action,Adventure,Fantasy
#> 6 6 The Great Wall Action,Adventure,Fantasy
#> 7 7 La La Land Comedy,Drama,Music
#> 8 8 Mindhorn Comedy
#> 9 9 The Lost City of Z Action,Adventure,Biography
#> 10 10 Passengers Adventure,Drama,Romance
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> 4 In a city of humanoid animals, a hustling theater impresario's attempt to save his theater with a singing competition becomes grander than he anticipates even as its finalists' find that their lives will never be the same.
#> 5 A secret government agency recruits some of the most dangerous incarcerated super-villains to form a defensive task force. Their first mission: save the world from the apocalypse.
#> 6 European mercenaries searching for black powder become embroiled in the defense of the Great Wall of China against a horde of monstrous creatures.
#> 7 A jazz pianist falls for an aspiring actress in Los Angeles.
#> 8 A has-been actor best known for playing the title character in the 1980s detective series "Mindhorn" must work with the police when a serial killer says that he will only speak with Detective Mindhorn, whom he believes to be a real person.
#> 9 A true-life drama, centering on British explorer Col. Percival Fawcett, who disappeared while searching for a mysterious city in the Amazon in the 1920s.
#> 10 A spacecraft traveling to a distant colony planet and transporting thousands of people has a malfunction in its sleep chambers. As a result, two passengers are awakened 90 years early.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> 4 Christophe Lourdelet
#> 5 David Ayer
#> 6 Yimou Zhang
#> 7 Damien Chazelle
#> 8 Sean Foley
#> 9 James Gray
#> 10 Morten Tyldum
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> 4 Matthew McConaughey,Reese Witherspoon, Seth MacFarlane, Scarlett Johansson
#> 5 Will Smith, Jared Leto, Margot Robbie, Viola Davis
#> 6 Matt Damon, Tian Jing, Willem Dafoe, Andy Lau
#> 7 Ryan Gosling, Emma Stone, Rosemarie DeWitt, J.K. Simmons
#> 8 Essie Davis, Andrea Riseborough, Julian Barratt,Kenneth Branagh
#> 9 Charlie Hunnam, Robert Pattinson, Sienna Miller, Tom Holland
#> 10 Jennifer Lawrence, Chris Pratt, Michael Sheen,Laurence Fishburne
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> 4 2016 108 7.2 60545 270.32
#> 5 2016 123 6.2 393727 325.02
#> 6 2016 103 6.1 56036 45.13
#> 7 2016 128 8.3 258682 151.06
#> 8 2016 89 6.4 2490 8.01
#> 9 2016 141 7.1 7188 8.01
#> 10 2016 116 7.0 192177 100.01
#> Metascore
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 42
#> 7 93
#> 8 71
#> 9 78
#> 10 41
fill(movies, RevenueMillions:Metascore, .direction = "up") %>%
head()
#> Rank Title Genre
#> 1 1 Guardians of the Galaxy Action,Adventure,Sci-Fi
#> 2 2 Prometheus Adventure,Mystery,Sci-Fi
#> 3 3 Split Horror,Thriller
#> 4 4 Sing Animation,Comedy,Family
#> 5 5 Suicide Squad Action,Adventure,Fantasy
#> 6 6 The Great Wall Action,Adventure,Fantasy
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> 4 In a city of humanoid animals, a hustling theater impresario's attempt to save his theater with a singing competition becomes grander than he anticipates even as its finalists' find that their lives will never be the same.
#> 5 A secret government agency recruits some of the most dangerous incarcerated super-villains to form a defensive task force. Their first mission: save the world from the apocalypse.
#> 6 European mercenaries searching for black powder become embroiled in the defense of the Great Wall of China against a horde of monstrous creatures.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> 4 Christophe Lourdelet
#> 5 David Ayer
#> 6 Yimou Zhang
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> 4 Matthew McConaughey,Reese Witherspoon, Seth MacFarlane, Scarlett Johansson
#> 5 Will Smith, Jared Leto, Margot Robbie, Viola Davis
#> 6 Matt Damon, Tian Jing, Willem Dafoe, Andy Lau
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> 4 2016 108 7.2 60545 270.32
#> 5 2016 123 6.2 393727 325.02
#> 6 2016 103 6.1 56036 45.13
#> Metascore
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 425.12.3 Replacing NA values by a constant
The function replace_na() replaces NA values with a constant value. It requires a named list of column names and values to replace NA values with. Pass in empty strings for the columns not to be affected.
# creating a named list of column values
lst <- list('','', 200, 50)
names(lst) <- names(movies)[1:3]
lst
#> $Rank
#> [1] ""
#>
#> $Title
#> [1] ""
#>
#> $Genre
#> [1] 200
#>
#> $<NA>
#> [1] 50
# replacing NA values with the named list
replace_na(movies, lst) %>%
head(10)
#> Rank Title Genre
#> 1 1 Guardians of the Galaxy Action,Adventure,Sci-Fi
#> 2 2 Prometheus Adventure,Mystery,Sci-Fi
#> 3 3 Split Horror,Thriller
#> 4 4 Sing Animation,Comedy,Family
#> 5 5 Suicide Squad Action,Adventure,Fantasy
#> 6 6 The Great Wall Action,Adventure,Fantasy
#> 7 7 La La Land Comedy,Drama,Music
#> 8 8 Mindhorn Comedy
#> 9 9 The Lost City of Z Action,Adventure,Biography
#> 10 10 Passengers Adventure,Drama,Romance
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> 4 In a city of humanoid animals, a hustling theater impresario's attempt to save his theater with a singing competition becomes grander than he anticipates even as its finalists' find that their lives will never be the same.
#> 5 A secret government agency recruits some of the most dangerous incarcerated super-villains to form a defensive task force. Their first mission: save the world from the apocalypse.
#> 6 European mercenaries searching for black powder become embroiled in the defense of the Great Wall of China against a horde of monstrous creatures.
#> 7 A jazz pianist falls for an aspiring actress in Los Angeles.
#> 8 A has-been actor best known for playing the title character in the 1980s detective series "Mindhorn" must work with the police when a serial killer says that he will only speak with Detective Mindhorn, whom he believes to be a real person.
#> 9 A true-life drama, centering on British explorer Col. Percival Fawcett, who disappeared while searching for a mysterious city in the Amazon in the 1920s.
#> 10 A spacecraft traveling to a distant colony planet and transporting thousands of people has a malfunction in its sleep chambers. As a result, two passengers are awakened 90 years early.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> 4 Christophe Lourdelet
#> 5 David Ayer
#> 6 Yimou Zhang
#> 7 Damien Chazelle
#> 8 Sean Foley
#> 9 James Gray
#> 10 Morten Tyldum
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> 4 Matthew McConaughey,Reese Witherspoon, Seth MacFarlane, Scarlett Johansson
#> 5 Will Smith, Jared Leto, Margot Robbie, Viola Davis
#> 6 Matt Damon, Tian Jing, Willem Dafoe, Andy Lau
#> 7 Ryan Gosling, Emma Stone, Rosemarie DeWitt, J.K. Simmons
#> 8 Essie Davis, Andrea Riseborough, Julian Barratt,Kenneth Branagh
#> 9 Charlie Hunnam, Robert Pattinson, Sienna Miller, Tom Holland
#> 10 Jennifer Lawrence, Chris Pratt, Michael Sheen,Laurence Fishburne
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> 4 2016 108 7.2 60545 270.32
#> 5 2016 123 6.2 393727 325.02
#> 6 2016 103 6.1 56036 45.13
#> 7 2016 128 8.3 258682 151.06
#> 8 2016 89 6.4 2490 NA
#> 9 2016 141 7.1 7188 8.01
#> 10 2016 116 7.0 192177 100.01
#> Metascore
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 42
#> 7 93
#> 8 71
#> 9 78
#> 10 41
# creating named list of computed values
lst <- list('',
'',
round(median(movies$RevenueMillions, na.rm = T), 2),
round(mean(movies$Metascore, na.rm = T)))
names(lst) <- names(movies)[1:4]
lst
#> $Rank
#> [1] ""
#>
#> $Title
#> [1] ""
#>
#> $Genre
#> [1] 47.98
#>
#> $Description
#> [1] 59
# replacing NA values
replace_na(movies, lst) %>%
head(10)
#> Rank Title Genre
#> 1 1 Guardians of the Galaxy Action,Adventure,Sci-Fi
#> 2 2 Prometheus Adventure,Mystery,Sci-Fi
#> 3 3 Split Horror,Thriller
#> 4 4 Sing Animation,Comedy,Family
#> 5 5 Suicide Squad Action,Adventure,Fantasy
#> 6 6 The Great Wall Action,Adventure,Fantasy
#> 7 7 La La Land Comedy,Drama,Music
#> 8 8 Mindhorn Comedy
#> 9 9 The Lost City of Z Action,Adventure,Biography
#> 10 10 Passengers Adventure,Drama,Romance
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> 4 In a city of humanoid animals, a hustling theater impresario's attempt to save his theater with a singing competition becomes grander than he anticipates even as its finalists' find that their lives will never be the same.
#> 5 A secret government agency recruits some of the most dangerous incarcerated super-villains to form a defensive task force. Their first mission: save the world from the apocalypse.
#> 6 European mercenaries searching for black powder become embroiled in the defense of the Great Wall of China against a horde of monstrous creatures.
#> 7 A jazz pianist falls for an aspiring actress in Los Angeles.
#> 8 A has-been actor best known for playing the title character in the 1980s detective series "Mindhorn" must work with the police when a serial killer says that he will only speak with Detective Mindhorn, whom he believes to be a real person.
#> 9 A true-life drama, centering on British explorer Col. Percival Fawcett, who disappeared while searching for a mysterious city in the Amazon in the 1920s.
#> 10 A spacecraft traveling to a distant colony planet and transporting thousands of people has a malfunction in its sleep chambers. As a result, two passengers are awakened 90 years early.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> 4 Christophe Lourdelet
#> 5 David Ayer
#> 6 Yimou Zhang
#> 7 Damien Chazelle
#> 8 Sean Foley
#> 9 James Gray
#> 10 Morten Tyldum
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> 4 Matthew McConaughey,Reese Witherspoon, Seth MacFarlane, Scarlett Johansson
#> 5 Will Smith, Jared Leto, Margot Robbie, Viola Davis
#> 6 Matt Damon, Tian Jing, Willem Dafoe, Andy Lau
#> 7 Ryan Gosling, Emma Stone, Rosemarie DeWitt, J.K. Simmons
#> 8 Essie Davis, Andrea Riseborough, Julian Barratt,Kenneth Branagh
#> 9 Charlie Hunnam, Robert Pattinson, Sienna Miller, Tom Holland
#> 10 Jennifer Lawrence, Chris Pratt, Michael Sheen,Laurence Fishburne
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> 4 2016 108 7.2 60545 270.32
#> 5 2016 123 6.2 393727 325.02
#> 6 2016 103 6.1 56036 45.13
#> 7 2016 128 8.3 258682 151.06
#> 8 2016 89 6.4 2490 NA
#> 9 2016 141 7.1 7188 8.01
#> 10 2016 116 7.0 192177 100.01
#> Metascore
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 42
#> 7 93
#> 8 71
#> 9 78
#> 10 415.12.4 Replacing NA values by groups
# splitting data frame
movies_split <- base::split(movies, movies$Year)
# replacing NA values
lapply(movies_split, function(x) {
lst <- list('',
'',
round(median(x[x$RevenueMillions, 'RevenueMillions'], na.rm = T),2),
round(mean(x[x$Metascore, 'Metascore'], na.rm = T)))
names(lst) <- names(movies)[1:4]
x <- replace_na(x, lst)
return(x)
}) %>%
dplyr::bind_rows() %>%
tail(10)
#> Rank Title
#> 991 948 King Cobra
#> 992 950 Kicks
#> 993 965 Custody
#> 994 967 L'odyssée
#> 995 975 Queen of Katwe
#> 996 976 My Big Fat Greek Wedding 2
#> 997 978 Amateur Night
#> 998 979 It's Only the End of the World
#> 999 981 Miracles from Heaven
#> 1000 1000 Nine Lives
#> Genre
#> 991 Crime,Drama
#> 992 Adventure
#> 993 Drama
#> 994 Adventure,Biography
#> 995 Biography,Drama,Sport
#> 996 Comedy,Family,Romance
#> 997 Comedy
#> 998 Drama
#> 999 Biography,Drama,Family
#> 1000 Comedy,Family,Fantasy
#> Description
#> 991 This ripped-from-the-headlines drama covers the early rise of gay porn headliner Sean Paul Lockhart a.k.a. Brent Corrigan, before his falling out with the producer who made him famous. When... See full summary »
#> 992 Brandon is a 15 year old whose dream is a pair of fresh Air Jordans. Soon after he gets his hands on them, they're stolen by a local hood, causing Brandon and his two friends to go on a dangerous mission through Oakland to retrieve them.
#> 993 The lives of three women are unexpectedly changed when they cross paths at a New York Family Court.
#> 994 Highly influential and a fearlessly ambitious pioneer, innovator, filmmaker, researcher and conservationist, Jacques-Yves Cousteau's aquatic adventure covers roughly thirty years of an inarguably rich in achievements life.
#> 995 A Ugandan girl sees her world rapidly change after being introduced to the game of chess.
#> 996 A Portokalos family secret brings the beloved characters back together for an even bigger and Greeker wedding.
#> 997 Guy Carter is an award-winning graduate student of architecture. He's got a beautiful wife and a baby on the way. The problem? He doesn't have "his ducks in a row," which only fuels his ... See full summary »
#> 998 Louis (Gaspard Ulliel), a terminally ill writer, returns home after a long absence to tell his family that he is dying.
#> 999 A young girl suffering from a rare digestive disorder finds herself miraculously cured after surviving a terrible accident.
#> 1000 A stuffy businessman finds himself trapped inside the body of his family's cat.
#> Director
#> 991 Justin Kelly
#> 992 Justin Tipping
#> 993 James Lapine
#> 994 Jérôme Salle
#> 995 Mira Nair
#> 996 Kirk Jones
#> 997 Lisa Addario
#> 998 Xavier Dolan
#> 999 Patricia Riggen
#> 1000 Barry Sonnenfeld
#> Actors
#> 991 Garrett Clayton, Christian Slater, Molly Ringwald,James Kelley
#> 992 Jahking Guillory, Christopher Jordan Wallace,Christopher Meyer, Kofi Siriboe
#> 993 Viola Davis, Hayden Panettiere, Catalina Sandino Moreno, Ellen Burstyn
#> 994 Lambert Wilson, Pierre Niney, Audrey Tautou,Laurent Lucas
#> 995 Madina Nalwanga, David Oyelowo, Lupita Nyong'o, Martin Kabanza
#> 996 Nia Vardalos, John Corbett, Michael Constantine, Lainie Kazan
#> 997 Jason Biggs, Janet Montgomery,Ashley Tisdale, Bria L. Murphy
#> 998 Nathalie Baye, Vincent Cassel, Marion Cotillard, Léa Seydoux
#> 999 Jennifer Garner, Kylie Rogers, Martin Henderson,Brighton Sharbino
#> 1000 Kevin Spacey, Jennifer Garner, Robbie Amell,Cheryl Hines
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 991 2016 91 5.6 3990 0.03
#> 992 2016 80 6.1 2417 0.15
#> 993 2016 104 6.9 280 NA
#> 994 2016 122 6.7 1810 NA
#> 995 2016 124 7.4 6753 8.81
#> 996 2016 94 6.0 20966 59.57
#> 997 2016 92 5.0 2229 NA
#> 998 2016 97 7.0 10658 NA
#> 999 2016 109 7.0 12048 61.69
#> 1000 2016 87 5.3 12435 19.64
#> Metascore
#> 991 48
#> 992 69
#> 993 72
#> 994 70
#> 995 73
#> 996 37
#> 997 38
#> 998 48
#> 999 44
#> 1000 115.12.5 Dropping NA values
The function drop_na() drops all rows containing NA values.
drop_na(movies) %>%
head(10)
#> Rank Title
#> 1 1 Guardians of the Galaxy
#> 2 2 Prometheus
#> 3 3 Split
#> 4 4 Sing
#> 5 5 Suicide Squad
#> 6 6 The Great Wall
#> 7 7 La La Land
#> 8 9 The Lost City of Z
#> 9 10 Passengers
#> 10 11 Fantastic Beasts and Where to Find Them
#> Genre
#> 1 Action,Adventure,Sci-Fi
#> 2 Adventure,Mystery,Sci-Fi
#> 3 Horror,Thriller
#> 4 Animation,Comedy,Family
#> 5 Action,Adventure,Fantasy
#> 6 Action,Adventure,Fantasy
#> 7 Comedy,Drama,Music
#> 8 Action,Adventure,Biography
#> 9 Adventure,Drama,Romance
#> 10 Adventure,Family,Fantasy
#> Description
#> 1 A group of intergalactic criminals are forced to work together to stop a fanatical warrior from taking control of the universe.
#> 2 Following clues to the origin of mankind, a team finds a structure on a distant moon, but they soon realize they are not alone.
#> 3 Three girls are kidnapped by a man with a diagnosed 23 distinct personalities. They must try to escape before the apparent emergence of a frightful new 24th.
#> 4 In a city of humanoid animals, a hustling theater impresario's attempt to save his theater with a singing competition becomes grander than he anticipates even as its finalists' find that their lives will never be the same.
#> 5 A secret government agency recruits some of the most dangerous incarcerated super-villains to form a defensive task force. Their first mission: save the world from the apocalypse.
#> 6 European mercenaries searching for black powder become embroiled in the defense of the Great Wall of China against a horde of monstrous creatures.
#> 7 A jazz pianist falls for an aspiring actress in Los Angeles.
#> 8 A true-life drama, centering on British explorer Col. Percival Fawcett, who disappeared while searching for a mysterious city in the Amazon in the 1920s.
#> 9 A spacecraft traveling to a distant colony planet and transporting thousands of people has a malfunction in its sleep chambers. As a result, two passengers are awakened 90 years early.
#> 10 The adventures of writer Newt Scamander in New York's secret community of witches and wizards seventy years before Harry Potter reads his book in school.
#> Director
#> 1 James Gunn
#> 2 Ridley Scott
#> 3 M. Night Shyamalan
#> 4 Christophe Lourdelet
#> 5 David Ayer
#> 6 Yimou Zhang
#> 7 Damien Chazelle
#> 8 James Gray
#> 9 Morten Tyldum
#> 10 David Yates
#> Actors
#> 1 Chris Pratt, Vin Diesel, Bradley Cooper, Zoe Saldana
#> 2 Noomi Rapace, Logan Marshall-Green, Michael Fassbender, Charlize Theron
#> 3 James McAvoy, Anya Taylor-Joy, Haley Lu Richardson, Jessica Sula
#> 4 Matthew McConaughey,Reese Witherspoon, Seth MacFarlane, Scarlett Johansson
#> 5 Will Smith, Jared Leto, Margot Robbie, Viola Davis
#> 6 Matt Damon, Tian Jing, Willem Dafoe, Andy Lau
#> 7 Ryan Gosling, Emma Stone, Rosemarie DeWitt, J.K. Simmons
#> 8 Charlie Hunnam, Robert Pattinson, Sienna Miller, Tom Holland
#> 9 Jennifer Lawrence, Chris Pratt, Michael Sheen,Laurence Fishburne
#> 10 Eddie Redmayne, Katherine Waterston, Alison Sudol,Dan Fogler
#> Year Runtime..Minutes. Rating Votes RevenueMillions
#> 1 2014 121 8.1 757074 333.13
#> 2 2012 124 7.0 485820 126.46
#> 3 2016 117 7.3 157606 138.12
#> 4 2016 108 7.2 60545 270.32
#> 5 2016 123 6.2 393727 325.02
#> 6 2016 103 6.1 56036 45.13
#> 7 2016 128 8.3 258682 151.06
#> 8 2016 141 7.1 7188 8.01
#> 9 2016 116 7.0 192177 100.01
#> 10 2016 133 7.5 232072 234.02
#> Metascore
#> 1 76
#> 2 65
#> 3 62
#> 4 59
#> 5 40
#> 6 42
#> 7 93
#> 8 78
#> 9 41
#> 10 66
drop_na(movies) %>%
nrow()
#> [1] 8385.13 Outliers
5.13.1 What is an outlier?
Outliers also known as anomalies are values that deviate extremely from other values within the same group of data. They occur because of errors committed while collecting or recording data, performing calculations or are just data points with extreme values.
5.13.2 Identifying outlier
5.13.2.1 Using summary statistics
The first step in outlier detection is to look at summary statistics, most especially the minimum, maximum, median, and mean. For example, with a dataset of people’s ages, if the maximum is 200 or the minimum is negative, then there is a problem.
library(gapminder)
data(gapminder)
gapminder_2007 <- subset(gapminder, year == '2007', select = -year)
head(gapminder_2007)
#> # A tibble: 6 x 5
#> country continent lifeExp pop gdpPercap
#> <fct> <fct> <dbl> <int> <dbl>
#> 1 Afghanistan Asia 43.8 31889923 975.
#> 2 Albania Europe 76.4 3600523 5937.
#> 3 Algeria Africa 72.3 33333216 6223.
#> 4 Angola Africa 42.7 12420476 4797.
#> 5 Argentina Americas 75.3 40301927 12779.
#> 6 Australia Oceania 81.2 20434176 34435.
summary(gapminder_2007$pop/1e6)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.1996 4.5080 10.5175 44.0212 31.2100 1318.6831From the above, we see that the median and mean are 10 million and 44 million respectively while the maximum value is 1.3 billion. This tells us that there are some outliers since the maximum value varies greatly from the centre of the data.
5.13.3 Using plots
Outliers are identified using univariate plots such as histogram, density plot and boxplot.
# plotting variable using histogram
hist(gapminder_2007$gdpPercap, breaks = 18)
# boxplot of population
boxplot(gapminder_2007$gdpPercap)
Of the above data visualizations, the boxplot is the most relevant as it shows both the spread of data and outliers. The boxplot reveals the following:
- minimum value,
- first quantile (Q1),
- median (second quantile),
- third quantile (Q3),
- maximum value excluding outliers and
- outliers.
The difference between Q3 and Q1 is known as the Interquartile Range (IQR).
The outliers within the box plot are calculated as any value that falls beyond 1.5 * IQR.
The function boxplot.stats() computes the data that is used to draw the box plot. Using this function, we can get our outliers.
boxplot.stats(gapminder_2007$gdpPercap)
#> $stats
#> [1] 277.5519 1598.4351 6124.3711 18008.9444 40675.9964
#>
#> $n
#> [1] 142
#>
#> $conf
#> [1] 3948.491 8300.251
#>
#> $out
#> [1] 47306.99 49357.19 47143.18 42951.65The first element returned is the summary statistic as was calculated with summary().
boxplot.stats(gapminder_2007$gdpPercap)$stats
#> [1] 277.5519 1598.4351 6124.3711 18008.9444 40675.9964
summary(gapminder_2007$gdpPercap)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 277.6 1624.8 6124.4 11680.1 18008.8 49357.2The last element returned are the outliers.
boxplot.stats(gapminder_2007$gdpPercap)$out
#> [1] 47306.99 49357.19 47143.18 42951.65Recall outliers are calculated as 1.5 * IQR, this can be changed using the argument coef. By default, it is set to 1.5 but can be changed as need be.
# changing coef
boxplot.stats(gapminder_2007$gdpPercap, coef = 0.8)$out
#> [1] 34435.37 36126.49 33692.61 36319.24 35278.42 33207.08
#> [7] 32170.37 39724.98 36180.79 40676.00 31656.07 47306.99
#> [13] 36797.93 49357.19 47143.18 33859.75 37506.42 33203.26
#> [19] 42951.65
boxplot.stats(gapminder_2007$gdpPercap, coef = 1)$out
#> [1] 34435.37 36126.49 36319.24 35278.42 39724.98 36180.79
#> [7] 40676.00 47306.99 36797.93 49357.19 47143.18 37506.42
#> [13] 42951.65
boxplot.stats(gapminder_2007$gdpPercap, coef = 1.2)$out
#> [1] 39724.98 40676.00 47306.99 49357.19 47143.18 42951.65
# selecting outliers
subset(gapminder_2007, gdpPercap >= min(boxplot.stats(gdpPercap)$out))
#> # A tibble: 4 x 5
#> country continent lifeExp pop gdpPercap
#> <fct> <fct> <dbl> <int> <dbl>
#> 1 Kuwait Asia 77.6 2505559 47307.
#> 2 Norway Europe 80.2 4627926 49357.
#> 3 Singapore Asia 80.0 4553009 47143.
#> 4 United States Americas 78.2 301139947 42952.5.13.4 Outliers by groups
# boxplot by continent
boxplot(gdpPercap ~ continent, gapminder_2007)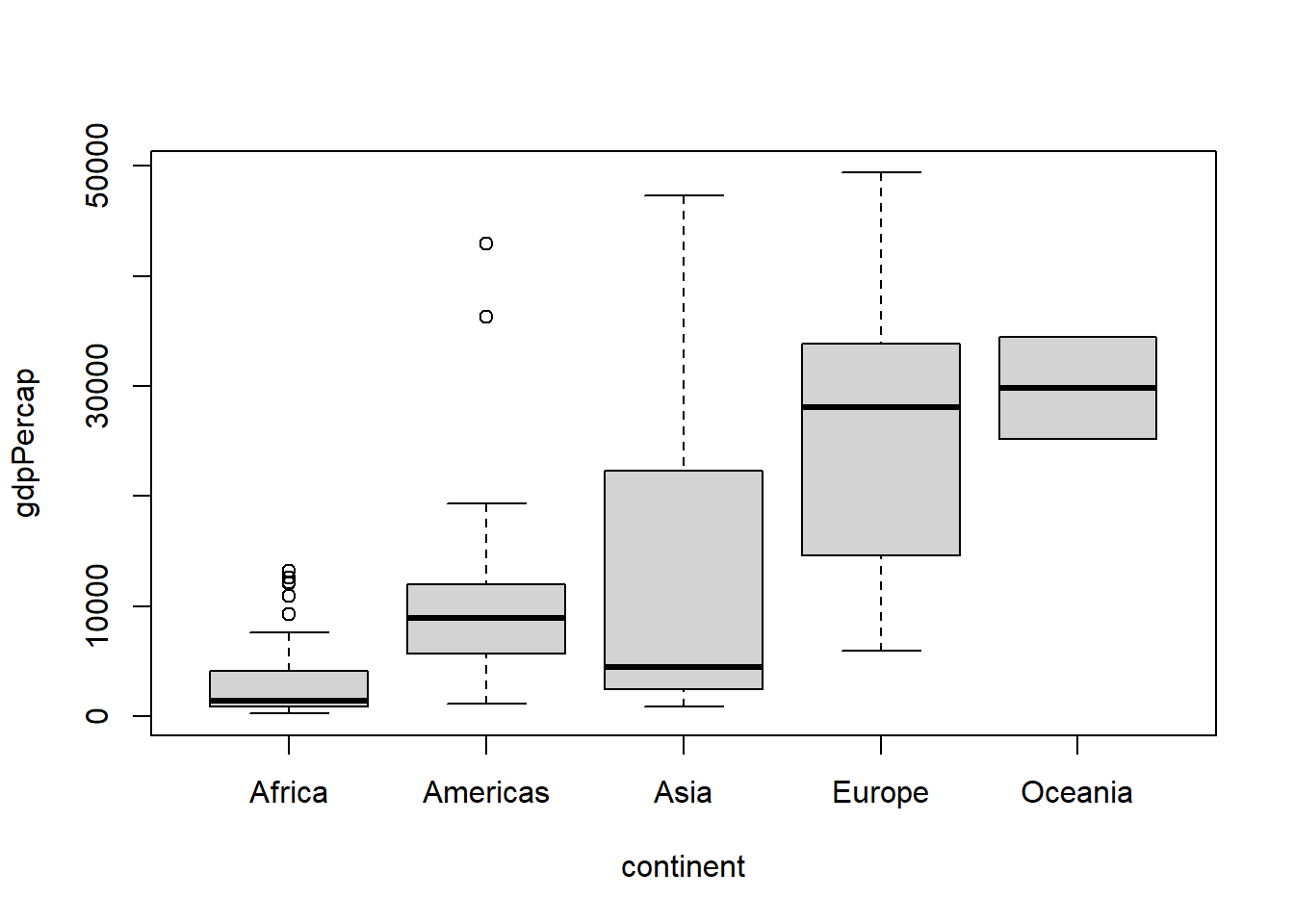
# splitting data frame
gap_split <- split(gapminder_2007, gapminder_2007$continent)
outliers_2007 <-
lapply(gap_split, function(x) {
x <- boxplot.stats(x$gdpPercap)$out
return(x)
})
outliers_2007
#> $Africa
#> [1] 12569.852 12154.090 13206.485 12057.499 10956.991
#> [6] 9269.658
#>
#> $Americas
#> [1] 36319.24 42951.65
#>
#> $Asia
#> numeric(0)
#>
#> $Europe
#> numeric(0)
#>
#> $Oceania
#> numeric(0)5.14 String manipulation with stringr
5.14.1 Determine string length
The function str_length() returns the count of letters in a string.
library(stringr)
month.name
#> [1] "January" "February" "March" "April"
#> [5] "May" "June" "July" "August"
#> [9] "September" "October" "November" "December"
str_length(month.name)
#> [1] 7 8 5 5 3 4 4 6 9 7 8 85.14.2 Strings formatting (case conversion)
The functions str_to_upper(), str_to_lower(), str_to_title() and str_to_sentence() are used to convert to upper, lower, title and sentence cases respectively.
The function str_pad() is used to pad characters before and/or after a string.
The function str_trunc() is used to truncate a string.
# lowercase
str_to_lower('It is an everyday thing', locale = "en")
#> [1] "it is an everyday thing"
# uppercase
str_to_upper('It is an everyday thing', locale = "en")
#> [1] "IT IS AN EVERYDAY THING"
# title case
str_to_title('It is an everyday thing', locale = "en")
#> [1] "It Is An Everyday Thing"
# sentence case
str_to_sentence('iT is aN everyDay thIng', locale = "en")
#> [1] "It is an everyday thing"
# padding string
str_pad(c(12, 235, 'abd', 'ame'), width = 5, pad = '0')
#> [1] "00012" "00235" "00abd" "00ame"
str_pad(c(12, 235, 'abd', 'ame'), width = 5, pad = 'X', side = 'right')
#> [1] "12XXX" "235XX" "abdXX" "ameXX"
str_pad(c(12, 235, 'abd', 'ame'), width = 5, pad = '-', side = 'both')
#> [1] "-12--" "-235-" "-abd-" "-ame-"
# truncate a character string
str_trunc(state.name[1:8], width = 6)
#> [1] "Ala..." "Alaska" "Ari..." "Ark..." "Cal..." "Col..."
#> [7] "Con..." "Del..."
str_trunc(state.name[1:8], 6, side = 'left')
#> [1] "...ama" "Alaska" "...ona" "...sas" "...nia" "...ado"
#> [7] "...cut" "...are"
str_trunc(state.name[1:8], 6, side = 'right', ellipsis = '')
#> [1] "Alabam" "Alaska" "Arizon" "Arkans" "Califo" "Colora"
#> [7] "Connec" "Delawa"5.14.3 Join and Split strings
5.14.3.1 joining strings with str_c()
The function str_c() joins two or more vectors element wise into a single character vector, optionally inserting separator (sep) between input vectors.
# combining elements into a character vector
str_c('a', 'b')
#> [1] "ab"
str_c(1, 2, 3, 4)
#> [1] "1234"
# using sep
str_c('a', 'b', sep = ' ')
#> [1] "a b"
str_c(1, 2, 3, 4, sep = ' ')
#> [1] "1 2 3 4"
str_c(1:10, sep = ' ')
#> [1] "1" "2" "3" "4" "5" "6" "7" "8" "9" "10"
# on a single vector
str_c(c('a', 'b'), sep = ' <> ')
#> [1] "a" "b"
str_c(c(1, 2), sep = ' <> ')
#> [1] "1" "2"
# two or more vectors
str_c(c('a', 'b'), c('c', 'd'), sep = ' <> ')
#> [1] "a <> c" "b <> d"
str_c(1:5, 10:20, sep = ' ')
#> [1] "1 10" "2 11" "3 12" "4 13" "5 14" "1 15" "2 16" "3 17"
#> [9] "4 18" "5 19" "1 20"
str_c(1:5, 10:20, c('a','b','c'), sep = ' ')
#> [1] "1 10 a" "2 11 b" "3 12 c" "4 13 a" "5 14 b" "1 15 c"
#> [7] "2 16 a" "3 17 b" "4 18 c" "5 19 a" "1 20 b"
# collapsing vectors
str_c(1:10, collapse = '~')
#> [1] "1~2~3~4~5~6~7~8~9~10"
str_c(c('a', 'b'), c('c', 'd'), collapse = ' <> ')
#> [1] "ac <> bd"
str_c(month.name[1:6], collapse = " - ")
#> [1] "January - February - March - April - May - June"
a <- month.name[1]
b <- month.name[2]
c <- month.name[3]
# combining character and variables
str_c(b,'comes after', a ,'but comes before', c, sep = " ")
#> [1] "February comes after January but comes before March"
str_c(b,'comes after', a ,'but comes before', c, sep = "/")
#> [1] "February/comes after/January/but comes before/March"
str_c('version 1.', 1:5, sep = '')
#> [1] "version 1.1" "version 1.2" "version 1.3" "version 1.4"
#> [5] "version 1.5"5.14.4 Joining using str_glue()
The function str_glue() returns a character vector containing a formatted combination of text and variable values.
formatting with integers
x <- 2
str_glue('{x} * {x} = {x ** 2}')
#> 2 * 2 = 4
x <- c(1:4)
str_glue('{x} squared is equal to {x ** 2}')
#> 1 squared is equal to 1
#> 2 squared is equal to 4
#> 3 squared is equal to 9
#> 4 squared is equal to 16
num <- c(123, 1, 100, 200, 10200, 25000)
str_glue('my registration number is {str_pad(num, 5, pad = "0")}')
#> my registration number is 00123
#> my registration number is 00001
#> my registration number is 00100
#> my registration number is 00200
#> my registration number is 10200
#> my registration number is 25000Formatting with strings
x <- 'my name is'
y <- 'james'
z <- 'london'
str_glue('{x} {y} and i live and work in {z}')
#> my name is james and i live and work in london
x <- 'my name is'
y <- 'james'
z <- 35
str_glue('{str_to_title(x)} {str_to_upper(y)} and i am {z} years')
#> My Name Is JAMES and i am 35 years
names <- c('paul', 'alphonse', 'michael', 'james', 'samson', 'terence', 'derin')
age <- c(30, 35, 32, 37, 29, 40, 30)
str_glue('i am {str_to_title(names)} and i am {age} years old')
#> i am Paul and i am 30 years old
#> i am Alphonse and i am 35 years old
#> i am Michael and i am 32 years old
#> i am James and i am 37 years old
#> i am Samson and i am 29 years old
#> i am Terence and i am 40 years old
#> i am Derin and i am 30 years oldFormatting with doubles or floating points
x <- 1000/6
x
#> [1] 166.6667
str_glue('1000 divided by 3 is {x}')
#> 1000 divided by 3 is 166.666666666667
str_glue('1000 divided by 3 is {round(x, 3)}')
#> 1000 divided by 3 is 166.667
str_glue('1000 divided by 3 is {round(x)}')
#> 1000 divided by 3 is 167
str_glue('1000 divided by 3 is {paste0("+", round(x))}')
#> 1000 divided by 3 is +167
str_glue('1000 divided by 3 is{paste0(" ", round(x))}')
#> 1000 divided by 3 is 1675.14.5 Splitting strings using str_split() and str_split_fixed()
The function str_split() splits the elements of a character vector into substrings by a specific pattern.
The function str_split_fixed() splits up the elements of a character into a fixed number of pieces.
str(str_split(c('2020-01-01', '2019-03-31', '2018-06-30'), pattern = "-"))
#> List of 3
#> $ : chr [1:3] "2020" "01" "01"
#> $ : chr [1:3] "2019" "03" "31"
#> $ : chr [1:3] "2018" "06" "30"
str(str_split(c('2020 01 01', '2019 03 31', '2018 06 30'), pattern = " "))
#> List of 3
#> $ : chr [1:3] "2020" "01" "01"
#> $ : chr [1:3] "2019" "03" "31"
#> $ : chr [1:3] "2018" "06" "30"
# splitting into two substrings
str(str_split(c('2020-01-01', '2019-03-31', '2018-06-30'), pattern = "-", n = 2))
#> List of 3
#> $ : chr [1:2] "2020" "01-01"
#> $ : chr [1:2] "2019" "03-31"
#> $ : chr [1:2] "2018" "06-30"
str(str_split(c('2020 01 01', '2019 03 31', '2018 06 30'), pattern = " ", n = 2))
#> List of 3
#> $ : chr [1:2] "2020" "01 01"
#> $ : chr [1:2] "2019" "03 31"
#> $ : chr [1:2] "2018" "06 30"
# returning a matrix
str_split_fixed(c('2020-01-01', '2019-03-31', '2018-06-30'), '-', 2)
#> [,1] [,2]
#> [1,] "2020" "01-01"
#> [2,] "2019" "03-31"
#> [3,] "2018" "06-30"
str_split_fixed(c('2020-01-01', '2019-03-31', '2018-06-30'), '-', 3)
#> [,1] [,2] [,3]
#> [1,] "2020" "01" "01"
#> [2,] "2019" "03" "31"
#> [3,] "2018" "06" "30"5.14.6 Extract and Replace part of a string
5.14.6.1 Extracting string values using str_sub()
The function str_sub() extracts a substring from a string by indexing. It uses start for the beginning position and stop for the ending position. It is like indexing but applied to a string.
var <- c('2020-01-01', '2019-03-31', '2018-06-30')
str_sub(var, start = 1, end = 4)
#> [1] "2020" "2019" "2018"
str_sub(var, 6, 7)
#> [1] "01" "03" "06"
str_sub(var, 9, 10)
#> [1] "01" "31" "30"
# using negative numbers
str_sub(var, -2, -1)
#> [1] "01" "31" "30"
str_sub(var, -5, -4)
#> [1] "01" "03" "06"
str_sub(var, -10, -7)
#> [1] "2020" "2019" "2018"5.14.6.2 Replacing string values using str_sub()
The function str_sub() is also used to replace substring in a string by assigning a different string to the extracted substring.
5.14.7 Replacing string values using str_replace()
The function str_replace() replaces a substring at first occurrence.
var <- c('2020-01-01', '2019-03-31', '2018-06-30')
str_replace(var, "-", "")
#> [1] "202001-01" "201903-31" "201806-30"
str_replace(var, "-", "/")
#> [1] "2020/01-01" "2019/03-31" "2018/06-30"5.14.8 Replacing string values using str_replace_all()
The function str_replace_all() replaces a substring throughout a string.
var <- c('2020-01-01', '2019-03-31', '2018-06-30')
str_replace_all(var, "-", " ")
#> [1] "2020 01 01" "2019 03 31" "2018 06 30"
str_replace_all(var, "-", "/")
#> [1] "2020/01/01" "2019/03/31" "2018/06/30"5.14.8.1 Remove white spaces and clean string values
The function:
-
str_trim()removes white spaces. -
str_squish()removes repeated spaces. -
str_remove()removes the first repeated spaces. -
str_remove_all()removes all repeated spaces.
# both sides
str_trim(c(' 2020-01-01 ', ' 2019-03-31 ', ' 2018-06-30 '))
#> [1] "2020-01-01" "2019-03-31" "2018-06-30"
# left side
str_trim(c(' 2020-01-01 ', ' 2019-03-31 ', ' 2018-06-30 '), side = 'left')
#> [1] "2020-01-01 " "2019-03-31 " "2018-06-30 "
# right side
str_trim(c(' 2020-01-01 ', ' 2019-03-31 ', ' 2018-06-30 '), side = 'right')
#> [1] " 2020-01-01" " 2019-03-31" " 2018-06-30"
str_squish('removing all repeated spaces in a string ')
#> [1] "removing all repeated spaces in a string"
str_remove('removing first repeated spaces in a string ', ' ')
#> [1] "removing first repeated spaces in a string "
str_remove_all('removing all repeated spaces in a string ', ' ')
#> [1] "removing allrepeated spaces in a string"5.14.9 Sorting
The function:
-
str_order()sorts a character vector and returns sorted indices. -
str_sort()sorts a character vector and returns sorted values.
str_order(month.name)
#> [1] 4 8 12 2 1 7 6 3 5 11 10 9
str_order(month.name, decreasing = T)
#> [1] 9 10 11 5 3 6 7 1 2 12 8 4
str_sort(month.name)
#> [1] "April" "August" "December" "February"
#> [5] "January" "July" "June" "March"
#> [9] "May" "November" "October" "September"
str_sort(month.name, decreasing = T)
#> [1] "September" "October" "November" "May"
#> [5] "March" "June" "July" "January"
#> [9] "February" "December" "August" "April"5.14.10 Duplicating strings
The function str_dup() duplicates and concatenate strings within a character vector.
5.14.11 Pattern matching using regular expression
5.14.11.1 Regex functions
-
str_which(),str_detect()andstr_subset()` str_count()-
str_starts()andstr_ends() -
str_locate()andstr_locate_all() -
str_extract()andstr_extract_all() -
str_match()andstr_match_all() -
str_view()andstr_view_all() -
str_replace()andstr_replace_all()
5.14.11.1.1 The functions str_detect(), str_which() and str_subset()
The function:
-
str_detect()detects the presence or absence of a pattern in a string and is equivalent to grepl(pattern, x). -
str_which()detects the position of a matched pattern and is equivalent to grep(pattern, x). -
str_subset()keeps string matching a pattern and is equivalent to grep(pattern, x, value = TRUE).
str_detect(month.name, 'uary')
#> [1] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [10] FALSE FALSE FALSE
month.name[str_detect(month.name, 'uary')]
#> [1] "January" "February"
str_detect(month.name, 'uary', negate = T)
#> [1] FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
#> [10] TRUE TRUE TRUE
month.name[str_detect(month.name, 'uary', negate = T)]
#> [1] "March" "April" "May" "June"
#> [5] "July" "August" "September" "October"
#> [9] "November" "December"
str_which(month.name, 'uary')
#> [1] 1 2
month.name[str_which(month.name, 'uary')]
#> [1] "January" "February"
str_which(month.name, 'uary', negate = T)
#> [1] 3 4 5 6 7 8 9 10 11 12
month.name[str_which(month.name, 'uary', negate = T)]
#> [1] "March" "April" "May" "June"
#> [5] "July" "August" "September" "October"
#> [9] "November" "December"
str_subset(month.name, pattern = 'ber')
#> [1] "September" "October" "November" "December"
str_subset(month.name, pattern = 'ber', negate = TRUE)
#> [1] "January" "February" "March" "April" "May"
#> [6] "June" "July" "August"5.14.11.1.2 The function str_count()
The function str_count() counts the number of matches in a string.
5.14.11.2 The functions str_starts() and str_ends()
The function:
-
str_starts()detects the presence of a pattern at the beginning of a string. -
str_ends()detects the presence of a pattern at the end of a string.
str_starts(month.name, 'J')
#> [1] TRUE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE
#> [10] FALSE FALSE FALSE
month.name[str_starts(month.name, 'J')]
#> [1] "January" "June" "July"
str_ends(month.name, 'ber')
#> [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE
#> [10] TRUE TRUE TRUE
month.name[str_ends(month.name, 'ber')]
#> [1] "September" "October" "November" "December"5.14.11.2.1 The functions str_locate() and str_locate_all()
The function:
-
str_locate()locates the position of the first pattern match in a string. -
str_locate_all()locates the position of all pattern matches in a string.
str_locate(month.name, 'ber')
#> start end
#> [1,] NA NA
#> [2,] NA NA
#> [3,] NA NA
#> [4,] NA NA
#> [5,] NA NA
#> [6,] NA NA
#> [7,] NA NA
#> [8,] NA NA
#> [9,] 7 9
#> [10,] 5 7
#> [11,] 6 8
#> [12,] 6 85.14.11.2.2 The functions str_extract() and str_extract_all()
The function:
-
str_extract()extracts the first matching pattern from a string. -
str_extract_all()extracts all matching patterns from a string.
str_extract(string = month.name, pattern = 'ber')
#> [1] NA NA NA NA NA NA NA NA "ber"
#> [10] "ber" "ber" "ber"5.14.11.2.3 The functions str_view() and str_view_all()
The functions str_view() and str_view_all() Views HTML rendering of regular expression match, with the first matching the first occurrence and the later all occurrences.
str_view(month.name, 'uary')5.14.11.3 Regex Operations
matching spaces
var <- c('2020 01 01', '2019 03 31', '2018 06 30')
str_replace_all(var, '[[:space:]]', '-')
#> [1] "2020-01-01" "2019-03-31" "2018-06-30"
str_replace_all(var, '\\s', '-')
#> [1] "2020-01-01" "2019-03-31" "2018-06-30"
str(str_split(var, '\\s'))
#> List of 3
#> $ : chr [1:3] "2020" "01" "01"
#> $ : chr [1:3] "2019" "03" "31"
#> $ : chr [1:3] "2018" "06" "30"
str_replace_all(var, '\\S', '-')
#> [1] "---- -- --" "---- -- --" "---- -- --"matching alphabetic characters
var <- 'a1b2c3d4e5f'
str_replace_all(var, '[[:alpha:]]', '')
#> [1] "12345"
# lowercase letters
str_replace_all(month.name, '[[:lower:]]', '')
#> [1] "J" "F" "M" "A" "M" "J" "J" "A" "S" "O" "N" "D"matching numerical digits
var <- 'a1b2c3d4e5f'
str_replace_all(var, '[[:digit:]]', '')
#> [1] "abcdef"
str_replace_all(var, '\\d', '')
#> [1] "abcdef"matching letters and numbers (alphanumeric characters)
var <- 'a1@; 2#4c $8`*%f^!1~0&^h*()j'
str_replace_all(var, '[[:alnum:]]', '')
#> [1] "@; # $`*%^!~&^*()"
str_replace_all(var, '[[:xdigit:]]', '')
#> [1] "@; # $`*%^!~&^h*()j"
str_replace_all(var, '\\w', '')
#> [1] "@; # $`*%^!~&^*()"matching punctuation
var <- 'a1@; 2#4c $8`*%f^!1~0&^h*()j'
str_replace_all(var, '[[:punct:]]', '')
#> [1] "a1 24c $8`f^1~0^hj"
str_replace_all(var, '\\W', '')
#> [1] "a124c8f10hj"matching letters, numbers, and punctuation
var <- 'a1@; 2#4c $8`*%f^!1~0&^h*()j'
str_replace_all(var, '[[:graph:]]', ' ')
#> [1] " "
str_replace_all(var, '.', ' ')
#> [1] " "matching whitespace
str_replace_all(c(' 2020-01-01 ', ' 2019-03-31 ', ' 2018-06-30 '), '\\s', '')
#> [1] "2020-01-01" "2019-03-31" "2018-06-30"matching newline and tap
cat('good morning \n i am fru kinglsy \n i will be your instructor')
#> good morning
#> i am fru kinglsy
#> i will be your instructor
# replacing new line
str_replace_all('good morning \n i am fru kinglsy \n i will be your instructor', '\\n', '\t')
#> [1] "good morning \t i am fru kinglsy \t i will be your instructor"
cat(str_replace_all('good morning \n i am fru kinglsy \n i will be your instructor', '\\n', '\t'))
#> good morning i am fru kinglsy i will be your instructor
# replacing tab
str_replace_all('good morning \t i am fru kinglsy \t i will be your instructor', '\\t', '\n')
#> [1] "good morning \n i am fru kinglsy \n i will be your instructor"
cat(str_replace_all('good morning \t i am fru kinglsy \t i will be your instructor', '\\t', '\n'))
#> good morning
#> i am fru kinglsy
#> i will be your instructormatching metacharacters
sales <- c('$25000', '$20000', '$22500', '$24000', '$30000', '$35000')
str_replace(sales, '\\$', '')
#> [1] "25000" "20000" "22500" "24000" "30000" "35000"
sales <- c('+25000', '+20000', '+22500', '+24000', '+30000', '+35000')
str_replace(sales, '\\+', '')
#> [1] "25000" "20000" "22500" "24000" "30000" "35000"
dates <- c('01.01.2012', '01.02.2012', '01.03.2012', '01.04.2012', '01.05.2012', '01.06.2012')
str_replace_all(dates, '\\.', '-')
#> [1] "01-01-2012" "01-02-2012" "01-03-2012" "01-04-2012"
#> [5] "01-05-2012" "01-06-2012"
dates <- c('01*01*2012', '01*02*2012', '01*03*2012', '01*04*2012', '01*05*2012', '01*06*2012')
str_replace_all(dates, '\\*', '-')
#> [1] "01-01-2012" "01-02-2012" "01-03-2012" "01-04-2012"
#> [5] "01-05-2012" "01-06-2012"
dates <- c('01^01^2012', '01^02^2012', '01^03^2012', '01^04^2012', '01^05^2012', '01^06^2012')
str_replace_all(dates, '\\^', '-')
#> [1] "01-01-2012" "01-02-2012" "01-03-2012" "01-04-2012"
#> [5] "01-05-2012" "01-06-2012"
dates <- c('01|01|2012', '01|02|2012', '01|03|2012', '01|04|2012', '01|05|2012', '01|06|2012')
str_replace_all(dates, '\\|', '-')
#> [1] "01-01-2012" "01-02-2012" "01-03-2012" "01-04-2012"
#> [5] "01-05-2012" "01-06-2012"
dates <- c('01\\01\\2012', '01\\02\\2012', '01\\03\\2012', '01\\04\\2012', '01\\05\\2012', '01\\06\\2012')
str_replace_all(dates, '\\\\', '-')
#> [1] "01-01-2012" "01-02-2012" "01-03-2012" "01-04-2012"
#> [5] "01-05-2012" "01-06-2012"
dates <- c('01\\.01\\.2012', '01\\.02\\.2012', '01\\.03\\.2012', '01\\.04\\.2012', '01\\.05\\.2012', '01\\.06\\.2012')
str_replace_all(dates, '\\\\\\.', '-')
#> [1] "01-01-2012" "01-02-2012" "01-03-2012" "01-04-2012"
#> [5] "01-05-2012" "01-06-2012"alternates and ranges
# either or
str_view_all(month.name, 'uary|ember|ober', '*')groups
str_subset(pattern = '(s{2})e', state.name)
#> [1] "Tennessee"anchors
# start of a string
str_replace_all(month.name, '^J', 'j')
#> [1] "january" "February" "March" "April"
#> [5] "May" "june" "july" "August"
#> [9] "September" "October" "November" "December"
# end of a string
str_replace_all(month.name, 'ber$', 'ba')
#> [1] "January" "February" "March" "April" "May"
#> [6] "June" "July" "August" "Septemba" "Octoba"
#> [11] "Novemba" "Decemba"quantifiers
# match 's' zero or one time
str_subset(month.name, 's?')
#> [1] "January" "February" "March" "April"
#> [5] "May" "June" "July" "August"
#> [9] "September" "October" "November" "December"
# match 'J' one or more times
str_subset(month.name, 'J+')
#> [1] "January" "June" "July"
# match 'e' one or more times
str_subset(state.name, 'e+')
#> [1] "Connecticut" "Delaware" "Georgia"
#> [4] "Kentucky" "Maine" "Massachusetts"
#> [7] "Minnesota" "Nebraska" "Nevada"
#> [10] "New Hampshire" "New Jersey" "New Mexico"
#> [13] "New York" "Oregon" "Pennsylvania"
#> [16] "Rhode Island" "Tennessee" "Texas"
#> [19] "Vermont" "West Virginia"
# matched 'y', zero or more times
str_subset(month.name, 'y*')
#> [1] "January" "February" "March" "April"
#> [5] "May" "June" "July" "August"
#> [9] "September" "October" "November" "December"
# matched 'a', zero or more times
str_subset(month.name, 'a*')
#> [1] "January" "February" "March" "April"
#> [5] "May" "June" "July" "August"
#> [9] "September" "October" "November" "December"
# match 'a' zero or more times and 'y'
str_subset(month.name, 'a*y')
#> [1] "January" "February" "May" "July"
# match 'y' zero or more times and 'a'
str_subset(month.name, 'y*a')
#> [1] "January" "February" "March" "May"
# match 's', exactly 2 times
str_subset(state.name, "s{2}")
#> [1] "Massachusetts" "Mississippi" "Missouri"
#> [4] "Tennessee"
# match 's', exactly 1 or more times
str_subset(state.name, "s{1,}")
#> [1] "Alaska" "Arkansas" "Illinois"
#> [4] "Kansas" "Louisiana" "Massachusetts"
#> [7] "Minnesota" "Mississippi" "Missouri"
#> [10] "Nebraska" "New Hampshire" "New Jersey"
#> [13] "Pennsylvania" "Rhode Island" "Tennessee"
#> [16] "Texas" "Washington" "West Virginia"
#> [19] "Wisconsin"
# match 's', exactly 1 or 2 times
str_subset(state.name, "s{1,2}")
#> [1] "Alaska" "Arkansas" "Illinois"
#> [4] "Kansas" "Louisiana" "Massachusetts"
#> [7] "Minnesota" "Mississippi" "Missouri"
#> [10] "Nebraska" "New Hampshire" "New Jersey"
#> [13] "Pennsylvania" "Rhode Island" "Tennessee"
#> [16] "Texas" "Washington" "West Virginia"
#> [19] "Wisconsin"